Gene
KWMTBOMO07868
Pre Gene Modal
BGIBMGA000949
Annotation
PREDICTED:_Bardet-Biedl_syndrome_7_protein_homolog_[Bombyx_mori]
Full name
Bardet-Biedl syndrome 7 protein homolog
Location in the cell
Extracellular Reliability : 1.304 Nuclear Reliability : 1.122
Sequence
CDS
ATGTCAGGAGCGACACGAGAGGGTTGGCAAGAGGTTGATATAACATCAGCAGTACCATTGGACATGCTGTTCATTTATTGTAGTAAGAAACTAAAATTACTGACTGACAGCGCTGCTGTCCTCAGCGTGTGCTCATCGCAGGATTCGGCGTCTGATTTGTTAGCAACTGTAAGATGTCAAGCAGGTACGAGGCGTATTTGGCTCCGTTTGCGTGTTGCATCAACTAATTCAACGGTTTTAGAGTCCACACGCGTATTAATATACGCATTACCGACAGGCGCGCCAAGAGTTGCAAGACTTTTATCAATAAAGCTACCAGCATTGCCGTACTATTCACAGCACGAGGAATTTGATGAAAATAATGACCACAGCATAAAGTCTTGGTGTCAACTCCACGTAACGGGTGGTTTTTCCGTTGCTGAAATGACTTTCTGGCTATCAGATATTTTACCCGGTGATTTACCGAGGCCTGCGTCAAATGTGGCTTTTTTACGTGTTCATTCTCTTTTGGGAACGAAATTATATTGCCAGTATCAACGAGGTTCAGCTGTATTTAAGTCAAACAATATTTCTACTGTATCATCAGTGAGAAGTATTATATCAAATTGTTCAATAGACAAAAGAATAAGAGTGGAAATATCGTGCGATATTCCTGATGATTGTTGTTTGAAATCATTCAAATTGATTGAAAACATCATTAGTATTGCATATGAAAAGAAAAACCAAGTCACTCTGGCGAAGGCTTTAGATTTACTAGAACTGGACACCAAGGCAATTAACGCCGAAAAACCTGTTTTATGTAAAGAATATTATACAGTTTTGAAAAATTCAGAAAGTGAAATATTACAATCAAATTTTGATGAACTCATAGATATTGTTGTACAATGGTACTTGGATTGGCGCTCATTATCTGTTAATGGTAACAACACAGTGGATCAATCAAATAAATTGTGTACCCTATTAAAAGACTATAAGCTTGATGATGTAACAAAAGTTCTTTCAGAACTAACAGAATTTAATCATACTTGTGAATAA
Protein
MSGATREGWQEVDITSAVPLDMLFIYCSKKLKLLTDSAAVLSVCSSQDSASDLLATVRCQAGTRRIWLRLRVASTNSTVLESTRVLIYALPTGAPRVARLLSIKLPALPYYSQHEEFDENNDHSIKSWCQLHVTGGFSVAEMTFWLSDILPGDLPRPASNVAFLRVHSLLGTKLYCQYQRGSAVFKSNNISTVSSVRSIISNCSIDKRIRVEISCDIPDDCCLKSFKLIENIISIAYEKKNQVTLAKALDLLELDTKAINAEKPVLCKEYYTVLKNSESEILQSNFDELIDIVVQWYLDWRSLSVNGNNTVDQSNKLCTLLKDYKLDDVTKVLSELTEFNHTCE
Summary
Description
The BBSome complex is thought to function as a coat complex required for sorting of specific membrane proteins to the primary cilia. The BBSome complex is required for ciliogenesis but is dispensable for centriolar satellite function.
Subunit
Part of BBSome complex.
Uniprot
A0A194RH81
A0A3S2LX33
A0A194Q979
A0A1B6CWW4
A0A2C9JHH1
E2C402
+ More
A0A2C9JHH6 A0A1D2MSA2 A0A0L7RAP5 A0A0N1ITH1 A0A2A3EEE7 A0A310SMG7 A0A0J7L2Z2 A0A088AMJ9 A0A154PBL4 N6TT07 E2B1J1 A0A0M0J419 A0A151X9E7 A0A2J7QS73 A0A3L8DUM0 A0A195BF52 A0A195FD92 A0A226EUV3 A0A067QWM4 A0A3S1HQB2 A0A210QW02 A0A1B0CA60 A0A151IMB3 E9IFJ9 A0A2R2MQG3 A0A2R2MQG4 A0A2R2MQF8 R7UUJ8 A0A1S3JU34 A0A1S3JTG6 A0A0B6ZN15 A0A026W9L5 A0A232ESC9 K7J2H3 A0A0L8GCJ1 A0A1X7VLG7 A0A0L8GCI8 A0A1V9X680 A0A3Q3ML98 A0A182WDY7 A0A1A7YSC2 F1Q6W8 Q08C18 A0A195DQX7 A0A023F648 A0A182MRP1 A0A1W4ZDB2 A0A0N8JWN9 C3XXL3 A0A369S8Z0 B3RXC9 V4B2U3 A0A1I8J8D3 A0A3R7MIN6 A0A2I4C6T3 A0A182K4S1 A0A3M6TJD7 A0A3Q2XSK6 D2A192 A0A1A8GTM9 A0A267FTV2 W4Z6L7 A0A1A8FKL1 A0A1A8R4F8 A0A1A8KHZ3 A0A1A8V4V8 A0A1A8C6M9 A0A1A8RDY3 A0A1A8LLG1 A0A1I7SS27 F6VNP5 A0A2B4SZC0 A0A3M7SJM9 A0A3B3WD55 A0A182Y9F8 A0A3Q2E4V6 A0A3P9NLI8 A0A182PP16 A0A3Q3FLG7 A0A3Q1IXQ6 A0A096LYQ5 A0A087YKI8 A0A3N0YML4 A0A3P9NLN8 A0A182L529
A0A2C9JHH6 A0A1D2MSA2 A0A0L7RAP5 A0A0N1ITH1 A0A2A3EEE7 A0A310SMG7 A0A0J7L2Z2 A0A088AMJ9 A0A154PBL4 N6TT07 E2B1J1 A0A0M0J419 A0A151X9E7 A0A2J7QS73 A0A3L8DUM0 A0A195BF52 A0A195FD92 A0A226EUV3 A0A067QWM4 A0A3S1HQB2 A0A210QW02 A0A1B0CA60 A0A151IMB3 E9IFJ9 A0A2R2MQG3 A0A2R2MQG4 A0A2R2MQF8 R7UUJ8 A0A1S3JU34 A0A1S3JTG6 A0A0B6ZN15 A0A026W9L5 A0A232ESC9 K7J2H3 A0A0L8GCJ1 A0A1X7VLG7 A0A0L8GCI8 A0A1V9X680 A0A3Q3ML98 A0A182WDY7 A0A1A7YSC2 F1Q6W8 Q08C18 A0A195DQX7 A0A023F648 A0A182MRP1 A0A1W4ZDB2 A0A0N8JWN9 C3XXL3 A0A369S8Z0 B3RXC9 V4B2U3 A0A1I8J8D3 A0A3R7MIN6 A0A2I4C6T3 A0A182K4S1 A0A3M6TJD7 A0A3Q2XSK6 D2A192 A0A1A8GTM9 A0A267FTV2 W4Z6L7 A0A1A8FKL1 A0A1A8R4F8 A0A1A8KHZ3 A0A1A8V4V8 A0A1A8C6M9 A0A1A8RDY3 A0A1A8LLG1 A0A1I7SS27 F6VNP5 A0A2B4SZC0 A0A3M7SJM9 A0A3B3WD55 A0A182Y9F8 A0A3Q2E4V6 A0A3P9NLI8 A0A182PP16 A0A3Q3FLG7 A0A3Q1IXQ6 A0A096LYQ5 A0A087YKI8 A0A3N0YML4 A0A3P9NLN8 A0A182L529
Pubmed
EMBL
KQ460205
KPJ16937.1
RSAL01000144
RVE45979.1
KQ459299
KPJ01974.1
+ More
GEDC01019306 GEDC01016748 JAS17992.1 JAS20550.1 GL452364 EFN77388.1 LJIJ01000599 ODM95967.1 KQ414618 KOC67903.1 KQ435798 KOX73408.1 KZ288280 PBC29662.1 KQ761208 OAD57989.1 LBMM01000942 KMQ97167.1 KQ434857 KZC08794.1 APGK01053107 APGK01053108 APGK01053109 APGK01053110 KB741221 ENN72415.1 GL444934 EFN60448.1 JWZX01003372 KOO21324.1 KQ982373 KYQ56997.1 NEVH01011878 PNF31420.1 QOIP01000004 RLU23992.1 KQ976500 KYM83198.1 KQ981673 KYN38351.1 LNIX01000002 OXA60998.1 KK852866 KDR14735.1 RQTK01000220 RUS84037.1 NEDP02001613 OWF52937.1 AJWK01003332 AJWK01003333 AJWK01003334 KQ977059 KYN06035.1 GL762841 EFZ20667.1 AMQN01007010 AMQN01007011 AMQN01007012 KB299759 ELU07577.1 HACG01023053 CEK69918.1 KK107372 EZA51709.1 NNAY01002440 OXU21269.1 KQ422516 KOF74746.1 KOF74747.1 MNPL01023042 OQR68893.1 HADW01004378 HADX01010864 SBP33096.1 BX004823 CABZ01077289 BC124452 AAI24453.1 KQ980581 KYN15278.1 GBBI01002029 JAC16683.1 AXCM01000907 JARO02009576 KPP61465.1 GG666471 EEN67473.1 NOWV01000054 RDD42483.1 DS985245 EDV24847.1 KB199753 ESP04393.1 NIVC01000382 PAA84302.1 QCYY01000659 ROT83651.1 RCHS01003480 RMX41547.1 KQ971338 EFA01565.1 HAEC01006249 SBQ74358.1 NIVC01000757 PAA77250.1 AAGJ04052172 AAGJ04052173 HAEB01013458 SBQ59985.1 HAEH01014730 SBS00861.1 HAED01008231 HAEE01011980 SBR32030.1 HADY01016527 HAEJ01015277 SBS55734.1 HADZ01010530 HAEA01010801 SBP74471.1 HAEI01008011 SBS03892.1 HAEF01007352 HAEG01013135 SBR44734.1 AAMC01120245 LSMT01000005 PFX34220.1 REGN01001244 RNA36084.1 AYCK01004184 RJVU01036043 ROL47160.1
GEDC01019306 GEDC01016748 JAS17992.1 JAS20550.1 GL452364 EFN77388.1 LJIJ01000599 ODM95967.1 KQ414618 KOC67903.1 KQ435798 KOX73408.1 KZ288280 PBC29662.1 KQ761208 OAD57989.1 LBMM01000942 KMQ97167.1 KQ434857 KZC08794.1 APGK01053107 APGK01053108 APGK01053109 APGK01053110 KB741221 ENN72415.1 GL444934 EFN60448.1 JWZX01003372 KOO21324.1 KQ982373 KYQ56997.1 NEVH01011878 PNF31420.1 QOIP01000004 RLU23992.1 KQ976500 KYM83198.1 KQ981673 KYN38351.1 LNIX01000002 OXA60998.1 KK852866 KDR14735.1 RQTK01000220 RUS84037.1 NEDP02001613 OWF52937.1 AJWK01003332 AJWK01003333 AJWK01003334 KQ977059 KYN06035.1 GL762841 EFZ20667.1 AMQN01007010 AMQN01007011 AMQN01007012 KB299759 ELU07577.1 HACG01023053 CEK69918.1 KK107372 EZA51709.1 NNAY01002440 OXU21269.1 KQ422516 KOF74746.1 KOF74747.1 MNPL01023042 OQR68893.1 HADW01004378 HADX01010864 SBP33096.1 BX004823 CABZ01077289 BC124452 AAI24453.1 KQ980581 KYN15278.1 GBBI01002029 JAC16683.1 AXCM01000907 JARO02009576 KPP61465.1 GG666471 EEN67473.1 NOWV01000054 RDD42483.1 DS985245 EDV24847.1 KB199753 ESP04393.1 NIVC01000382 PAA84302.1 QCYY01000659 ROT83651.1 RCHS01003480 RMX41547.1 KQ971338 EFA01565.1 HAEC01006249 SBQ74358.1 NIVC01000757 PAA77250.1 AAGJ04052172 AAGJ04052173 HAEB01013458 SBQ59985.1 HAEH01014730 SBS00861.1 HAED01008231 HAEE01011980 SBR32030.1 HADY01016527 HAEJ01015277 SBS55734.1 HADZ01010530 HAEA01010801 SBP74471.1 HAEI01008011 SBS03892.1 HAEF01007352 HAEG01013135 SBR44734.1 AAMC01120245 LSMT01000005 PFX34220.1 REGN01001244 RNA36084.1 AYCK01004184 RJVU01036043 ROL47160.1
Proteomes
UP000053240
UP000283053
UP000053268
UP000076420
UP000008237
UP000094527
+ More
UP000053825 UP000053105 UP000242457 UP000036403 UP000005203 UP000076502 UP000019118 UP000000311 UP000075809 UP000235965 UP000279307 UP000078540 UP000078541 UP000198287 UP000027135 UP000271974 UP000242188 UP000092461 UP000078542 UP000085678 UP000014760 UP000053097 UP000215335 UP000002358 UP000053454 UP000007879 UP000192247 UP000261640 UP000075920 UP000000437 UP000078492 UP000075883 UP000192224 UP000034805 UP000001554 UP000253843 UP000009022 UP000030746 UP000095280 UP000215902 UP000283509 UP000192220 UP000075881 UP000275408 UP000264820 UP000007266 UP000007110 UP000095284 UP000008143 UP000225706 UP000276133 UP000261480 UP000076408 UP000265020 UP000242638 UP000075885 UP000261660 UP000265040 UP000028760 UP000075882
UP000053825 UP000053105 UP000242457 UP000036403 UP000005203 UP000076502 UP000019118 UP000000311 UP000075809 UP000235965 UP000279307 UP000078540 UP000078541 UP000198287 UP000027135 UP000271974 UP000242188 UP000092461 UP000078542 UP000085678 UP000014760 UP000053097 UP000215335 UP000002358 UP000053454 UP000007879 UP000192247 UP000261640 UP000075920 UP000000437 UP000078492 UP000075883 UP000192224 UP000034805 UP000001554 UP000253843 UP000009022 UP000030746 UP000095280 UP000215902 UP000283509 UP000192220 UP000075881 UP000275408 UP000264820 UP000007266 UP000007110 UP000095284 UP000008143 UP000225706 UP000276133 UP000261480 UP000076408 UP000265020 UP000242638 UP000075885 UP000261660 UP000265040 UP000028760 UP000075882
Interpro
Gene 3D
ProteinModelPortal
A0A194RH81
A0A3S2LX33
A0A194Q979
A0A1B6CWW4
A0A2C9JHH1
E2C402
+ More
A0A2C9JHH6 A0A1D2MSA2 A0A0L7RAP5 A0A0N1ITH1 A0A2A3EEE7 A0A310SMG7 A0A0J7L2Z2 A0A088AMJ9 A0A154PBL4 N6TT07 E2B1J1 A0A0M0J419 A0A151X9E7 A0A2J7QS73 A0A3L8DUM0 A0A195BF52 A0A195FD92 A0A226EUV3 A0A067QWM4 A0A3S1HQB2 A0A210QW02 A0A1B0CA60 A0A151IMB3 E9IFJ9 A0A2R2MQG3 A0A2R2MQG4 A0A2R2MQF8 R7UUJ8 A0A1S3JU34 A0A1S3JTG6 A0A0B6ZN15 A0A026W9L5 A0A232ESC9 K7J2H3 A0A0L8GCJ1 A0A1X7VLG7 A0A0L8GCI8 A0A1V9X680 A0A3Q3ML98 A0A182WDY7 A0A1A7YSC2 F1Q6W8 Q08C18 A0A195DQX7 A0A023F648 A0A182MRP1 A0A1W4ZDB2 A0A0N8JWN9 C3XXL3 A0A369S8Z0 B3RXC9 V4B2U3 A0A1I8J8D3 A0A3R7MIN6 A0A2I4C6T3 A0A182K4S1 A0A3M6TJD7 A0A3Q2XSK6 D2A192 A0A1A8GTM9 A0A267FTV2 W4Z6L7 A0A1A8FKL1 A0A1A8R4F8 A0A1A8KHZ3 A0A1A8V4V8 A0A1A8C6M9 A0A1A8RDY3 A0A1A8LLG1 A0A1I7SS27 F6VNP5 A0A2B4SZC0 A0A3M7SJM9 A0A3B3WD55 A0A182Y9F8 A0A3Q2E4V6 A0A3P9NLI8 A0A182PP16 A0A3Q3FLG7 A0A3Q1IXQ6 A0A096LYQ5 A0A087YKI8 A0A3N0YML4 A0A3P9NLN8 A0A182L529
A0A2C9JHH6 A0A1D2MSA2 A0A0L7RAP5 A0A0N1ITH1 A0A2A3EEE7 A0A310SMG7 A0A0J7L2Z2 A0A088AMJ9 A0A154PBL4 N6TT07 E2B1J1 A0A0M0J419 A0A151X9E7 A0A2J7QS73 A0A3L8DUM0 A0A195BF52 A0A195FD92 A0A226EUV3 A0A067QWM4 A0A3S1HQB2 A0A210QW02 A0A1B0CA60 A0A151IMB3 E9IFJ9 A0A2R2MQG3 A0A2R2MQG4 A0A2R2MQF8 R7UUJ8 A0A1S3JU34 A0A1S3JTG6 A0A0B6ZN15 A0A026W9L5 A0A232ESC9 K7J2H3 A0A0L8GCJ1 A0A1X7VLG7 A0A0L8GCI8 A0A1V9X680 A0A3Q3ML98 A0A182WDY7 A0A1A7YSC2 F1Q6W8 Q08C18 A0A195DQX7 A0A023F648 A0A182MRP1 A0A1W4ZDB2 A0A0N8JWN9 C3XXL3 A0A369S8Z0 B3RXC9 V4B2U3 A0A1I8J8D3 A0A3R7MIN6 A0A2I4C6T3 A0A182K4S1 A0A3M6TJD7 A0A3Q2XSK6 D2A192 A0A1A8GTM9 A0A267FTV2 W4Z6L7 A0A1A8FKL1 A0A1A8R4F8 A0A1A8KHZ3 A0A1A8V4V8 A0A1A8C6M9 A0A1A8RDY3 A0A1A8LLG1 A0A1I7SS27 F6VNP5 A0A2B4SZC0 A0A3M7SJM9 A0A3B3WD55 A0A182Y9F8 A0A3Q2E4V6 A0A3P9NLI8 A0A182PP16 A0A3Q3FLG7 A0A3Q1IXQ6 A0A096LYQ5 A0A087YKI8 A0A3N0YML4 A0A3P9NLN8 A0A182L529
Ontologies
KEGG
GO
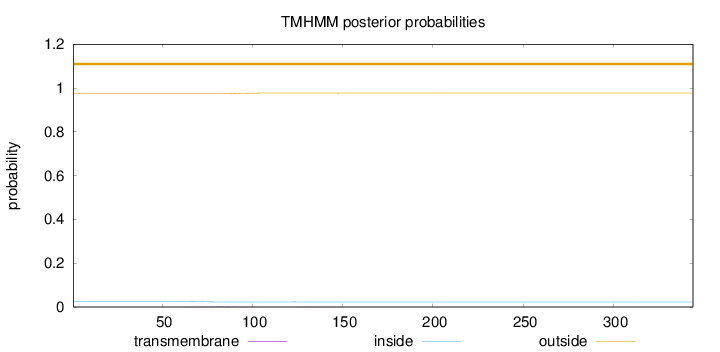
Topology
Length:
344
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0431800000000001
Exp number, first 60 AAs:
0.00227
Total prob of N-in:
0.02472
outside
1 - 344
Population Genetic Test Statistics
Pi
188.433733
Theta
228.291732
Tajima's D
-0.581289
CLR
0.776719
CSRT
0.221688915554222
Interpretation
Uncertain
