Gene
KWMTBOMO07867
Pre Gene Modal
BGIBMGA000949
Annotation
PREDICTED:_Bardet-Biedl_syndrome_7_protein_homolog_[Bombyx_mori]
Full name
Bardet-Biedl syndrome 7 protein homolog
Location in the cell
PlasmaMembrane Reliability : 3.305
Sequence
CDS
ATGTTAAAGATTTTATGTCAGCAGAGTGAAAAAGACGTTGCCGGTATTATTATCATATATTATAGAGTTATGGATTACGACTTGTGCAGAATTGACTACTTTATAAGCGGAATCACATATCCTGACACTCTAAAAATTTTACCATCTCAAGGACAGAAACTGGAACAGAAGGTCGTAGTTGGGGATAAAACCGGAGTTCTGCAATGTTTGACCATAAGAGACGAAGAACCAGTCATTCAATTTAAGACATTGCCAGGCAAACCAATTACCGCCGTACAATTGACTTCTACAAACGGTACGCAACCAGACAAAATATTTGCAGCATCGGGTAATGAGGTTAAAGGTTACACCAAAAAAGGCAAAGTATTCTTCTCTATTGAGACAGCATTGTCGGAGCCTGTGACGTCTATGTGTGTTTTCGGTGGTGATCTAATACTATGCAGTGGACGGACCATCACATTTTATCGTGATTCGAAGGAACATCAGTCATATGTTTGTAAAGATAGAGTTTTGGATATGGCAGCCTTTATTGCACCGAATAATGCTAAGGTGCGACTGCTTCTTTTGATTGCACATAAAGGCGCGGGCATATTAGAGAATGGACATCTGGTAGCTCACATAGCTATATCTTCTGGTCCCTCTCGATTAGCGACACCGCCTGTTGCTCAAGTTGCAGAAATTAAATGTTTCTATGGTGCGGCCGATGGTTCTGTGGGTATTATTACCTATGAAGAAACGACACTTTCAAGCAATTGTCTTGTAGAAGGCCGAGGCCTAGGTTCTGTAGTGTGCCTTGGATGGTTTTATTCTAATGCAAATCTCCACCTCGCTGTAGGGCGAAACGATGGATCTGTTCAGCTCTATCTCATAGACGTTGAGAATCTGCATCTCAAACCAAGACTCAAATTTACATATGTACGTATATGTATACAAATTATTAATATATATTTCCATTGA
Protein
MLKILCQQSEKDVAGIIIIYYRVMDYDLCRIDYFISGITYPDTLKILPSQGQKLEQKVVVGDKTGVLQCLTIRDEEPVIQFKTLPGKPITAVQLTSTNGTQPDKIFAASGNEVKGYTKKGKVFFSIETALSEPVTSMCVFGGDLILCSGRTITFYRDSKEHQSYVCKDRVLDMAAFIAPNNAKVRLLLLIAHKGAGILENGHLVAHIAISSGPSRLATPPVAQVAEIKCFYGAADGSVGIITYEETTLSSNCLVEGRGLGSVVCLGWFYSNANLHLAVGRNDGSVQLYLIDVENLHLKPRLKFTYVRICIQIINIYFH
Summary
Description
The BBSome complex is thought to function as a coat complex required for sorting of specific membrane proteins to the primary cilia. The BBSome complex is required for ciliogenesis but is dispensable for centriolar satellite function.
Subunit
Part of BBSome complex.
Uniprot
A0A2H1VXJ3
A0A3S2LX33
A0A0L7LAS6
A0A194Q979
A0A194RH81
A0A212F4G0
+ More
A0A1J1IQ23 D2A192 U4U670 N6TT07 A0A2J7QS43 A0A067QWM4 Q7PZG7 A0A182SBT9 A0A2J7QS73 A0A182QGI9 A0A182RS06 A0A182XJT9 A0A182UGI4 A0A182L529 A0A182VHN7 A0A182MRP1 A0A182G9L2 A0A182HHJ3 A0A3B3SW41 A0A182HB65 Q17MR4 A0A1S3RSF0 C0H9X4 V9KW78 A0A1W4ZDB2 A0A084WIN3 A0A1B0CA60 H2ZZ55 A0A2G8LLF3 A0A182N2V8 A0A3P8UZ40 V9KJA7 W4XPH5 A0A182FXJ9 A0A3P8X9T0 A0A3B4YF00 A0A3Q1HLH7 A0A3Q1FZW5 A0A3Q1IXQ6 A0A3Q3FLC8 A0A182PP16 W5M531 A0A3P8TRR5 A0A182Y9F8 A0A3Q2VZS3 A0A2D0RI98 E0VDR8 A0A2U9C684 I3IYL8 E1BXI4 A0A3Q1AVS2 E9QDJ5 A0A3P9B089 A0A3B4ZP69 A0A3M6TJD7 A0A1S3JTG6 A0A3Q4HCN3 A0A1S3JU34 A0A182WDY7 A0A3B4TYI6 A0A2B4SZC0 A0A2R2MQG4 A0A2R2MQG3 A0A2R2MQF8 A0A182J9S2 A0A0Q3X8Z7 A0A3Q2Z8G1 A0A3Q2XSK6 F1Q6W8 W5L0U3 A0A3Q3J8C4 A0A3B4DAQ5 A0A336LLY4 A0A3N0YML4 A0A336M1K3 H3D688 Q08C18 A0A3Q3MSC5 A0A3B3CZS6 A0A0K8T3A0 A0A1U8DDU9 T1IN54 A0A1U8D6D0 A0A1U7R903 A0A2G8JCI2 A0A3Q2E4V6 A0A182K4S1 F6XU90 G3PWM5 A0A023F648
A0A1J1IQ23 D2A192 U4U670 N6TT07 A0A2J7QS43 A0A067QWM4 Q7PZG7 A0A182SBT9 A0A2J7QS73 A0A182QGI9 A0A182RS06 A0A182XJT9 A0A182UGI4 A0A182L529 A0A182VHN7 A0A182MRP1 A0A182G9L2 A0A182HHJ3 A0A3B3SW41 A0A182HB65 Q17MR4 A0A1S3RSF0 C0H9X4 V9KW78 A0A1W4ZDB2 A0A084WIN3 A0A1B0CA60 H2ZZ55 A0A2G8LLF3 A0A182N2V8 A0A3P8UZ40 V9KJA7 W4XPH5 A0A182FXJ9 A0A3P8X9T0 A0A3B4YF00 A0A3Q1HLH7 A0A3Q1FZW5 A0A3Q1IXQ6 A0A3Q3FLC8 A0A182PP16 W5M531 A0A3P8TRR5 A0A182Y9F8 A0A3Q2VZS3 A0A2D0RI98 E0VDR8 A0A2U9C684 I3IYL8 E1BXI4 A0A3Q1AVS2 E9QDJ5 A0A3P9B089 A0A3B4ZP69 A0A3M6TJD7 A0A1S3JTG6 A0A3Q4HCN3 A0A1S3JU34 A0A182WDY7 A0A3B4TYI6 A0A2B4SZC0 A0A2R2MQG4 A0A2R2MQG3 A0A2R2MQF8 A0A182J9S2 A0A0Q3X8Z7 A0A3Q2Z8G1 A0A3Q2XSK6 F1Q6W8 W5L0U3 A0A3Q3J8C4 A0A3B4DAQ5 A0A336LLY4 A0A3N0YML4 A0A336M1K3 H3D688 Q08C18 A0A3Q3MSC5 A0A3B3CZS6 A0A0K8T3A0 A0A1U8DDU9 T1IN54 A0A1U8D6D0 A0A1U7R903 A0A2G8JCI2 A0A3Q2E4V6 A0A182K4S1 F6XU90 G3PWM5 A0A023F648
Pubmed
EMBL
ODYU01005045
SOQ45543.1
RSAL01000144
RVE45979.1
JTDY01001901
KOB72577.1
+ More
KQ459299 KPJ01974.1 KQ460205 KPJ16937.1 AGBW02010374 OWR48621.1 CVRI01000057 CRL02341.1 KQ971338 EFA01565.1 KB632081 ERL88577.1 APGK01053107 APGK01053108 APGK01053109 APGK01053110 KB741221 ENN72415.1 NEVH01011878 PNF31421.1 KK852866 KDR14735.1 AAAB01008986 EAA00146.3 PNF31420.1 AXCN02001090 AXCM01000907 JXUM01049375 KQ561601 KXJ78034.1 APCN01002417 JXUM01031757 KQ560934 KXJ80290.1 CH477204 EAT47958.1 BT059130 ACN10843.1 JW870798 AFP03316.1 ATLV01023941 KE525347 KFB50077.1 AJWK01003332 AJWK01003333 AJWK01003334 AFYH01181242 AFYH01181243 AFYH01181244 AFYH01181245 AFYH01181246 AFYH01181247 AFYH01181248 AFYH01181249 AFYH01181250 AFYH01181251 MRZV01000041 PIK61079.1 JW865561 AFO98078.1 AAGJ04052172 AHAT01020575 AHAT01020576 DS235083 EEB11524.1 CP026255 AWP11703.1 AERX01009930 AADN05000017 BX004823 CABZ01077289 RCHS01003480 RMX41547.1 LSMT01000005 PFX34220.1 LMAW01000230 KQL59941.1 UFQS01002849 UFQT01002849 SSX14742.1 SSX34134.1 RJVU01036043 ROL47160.1 UFQS01000416 UFQT01000416 SSX03749.1 SSX24114.1 BC124452 AAI24453.1 GBRD01006151 JAG59670.1 JH431145 MRZV01002551 PIK33452.1 AAPN01300085 AAPN01300086 GBBI01002029 JAC16683.1
KQ459299 KPJ01974.1 KQ460205 KPJ16937.1 AGBW02010374 OWR48621.1 CVRI01000057 CRL02341.1 KQ971338 EFA01565.1 KB632081 ERL88577.1 APGK01053107 APGK01053108 APGK01053109 APGK01053110 KB741221 ENN72415.1 NEVH01011878 PNF31421.1 KK852866 KDR14735.1 AAAB01008986 EAA00146.3 PNF31420.1 AXCN02001090 AXCM01000907 JXUM01049375 KQ561601 KXJ78034.1 APCN01002417 JXUM01031757 KQ560934 KXJ80290.1 CH477204 EAT47958.1 BT059130 ACN10843.1 JW870798 AFP03316.1 ATLV01023941 KE525347 KFB50077.1 AJWK01003332 AJWK01003333 AJWK01003334 AFYH01181242 AFYH01181243 AFYH01181244 AFYH01181245 AFYH01181246 AFYH01181247 AFYH01181248 AFYH01181249 AFYH01181250 AFYH01181251 MRZV01000041 PIK61079.1 JW865561 AFO98078.1 AAGJ04052172 AHAT01020575 AHAT01020576 DS235083 EEB11524.1 CP026255 AWP11703.1 AERX01009930 AADN05000017 BX004823 CABZ01077289 RCHS01003480 RMX41547.1 LSMT01000005 PFX34220.1 LMAW01000230 KQL59941.1 UFQS01002849 UFQT01002849 SSX14742.1 SSX34134.1 RJVU01036043 ROL47160.1 UFQS01000416 UFQT01000416 SSX03749.1 SSX24114.1 BC124452 AAI24453.1 GBRD01006151 JAG59670.1 JH431145 MRZV01002551 PIK33452.1 AAPN01300085 AAPN01300086 GBBI01002029 JAC16683.1
Proteomes
UP000283053
UP000037510
UP000053268
UP000053240
UP000007151
UP000183832
+ More
UP000007266 UP000030742 UP000019118 UP000235965 UP000027135 UP000007062 UP000075901 UP000075886 UP000075900 UP000076407 UP000075902 UP000075882 UP000075903 UP000075883 UP000069940 UP000249989 UP000075840 UP000261540 UP000008820 UP000087266 UP000192224 UP000030765 UP000092461 UP000008672 UP000230750 UP000075884 UP000265120 UP000007110 UP000069272 UP000265140 UP000261360 UP000265040 UP000257200 UP000261660 UP000075885 UP000018468 UP000265080 UP000076408 UP000264840 UP000221080 UP000009046 UP000246464 UP000005207 UP000000539 UP000257160 UP000000437 UP000265160 UP000261400 UP000275408 UP000085678 UP000261580 UP000075920 UP000261420 UP000225706 UP000075880 UP000051836 UP000264820 UP000018467 UP000261600 UP000261440 UP000007303 UP000261640 UP000261560 UP000189705 UP000265020 UP000075881 UP000002279 UP000007635
UP000007266 UP000030742 UP000019118 UP000235965 UP000027135 UP000007062 UP000075901 UP000075886 UP000075900 UP000076407 UP000075902 UP000075882 UP000075903 UP000075883 UP000069940 UP000249989 UP000075840 UP000261540 UP000008820 UP000087266 UP000192224 UP000030765 UP000092461 UP000008672 UP000230750 UP000075884 UP000265120 UP000007110 UP000069272 UP000265140 UP000261360 UP000265040 UP000257200 UP000261660 UP000075885 UP000018468 UP000265080 UP000076408 UP000264840 UP000221080 UP000009046 UP000246464 UP000005207 UP000000539 UP000257160 UP000000437 UP000265160 UP000261400 UP000275408 UP000085678 UP000261580 UP000075920 UP000261420 UP000225706 UP000075880 UP000051836 UP000264820 UP000018467 UP000261600 UP000261440 UP000007303 UP000261640 UP000261560 UP000189705 UP000265020 UP000075881 UP000002279 UP000007635
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VXJ3
A0A3S2LX33
A0A0L7LAS6
A0A194Q979
A0A194RH81
A0A212F4G0
+ More
A0A1J1IQ23 D2A192 U4U670 N6TT07 A0A2J7QS43 A0A067QWM4 Q7PZG7 A0A182SBT9 A0A2J7QS73 A0A182QGI9 A0A182RS06 A0A182XJT9 A0A182UGI4 A0A182L529 A0A182VHN7 A0A182MRP1 A0A182G9L2 A0A182HHJ3 A0A3B3SW41 A0A182HB65 Q17MR4 A0A1S3RSF0 C0H9X4 V9KW78 A0A1W4ZDB2 A0A084WIN3 A0A1B0CA60 H2ZZ55 A0A2G8LLF3 A0A182N2V8 A0A3P8UZ40 V9KJA7 W4XPH5 A0A182FXJ9 A0A3P8X9T0 A0A3B4YF00 A0A3Q1HLH7 A0A3Q1FZW5 A0A3Q1IXQ6 A0A3Q3FLC8 A0A182PP16 W5M531 A0A3P8TRR5 A0A182Y9F8 A0A3Q2VZS3 A0A2D0RI98 E0VDR8 A0A2U9C684 I3IYL8 E1BXI4 A0A3Q1AVS2 E9QDJ5 A0A3P9B089 A0A3B4ZP69 A0A3M6TJD7 A0A1S3JTG6 A0A3Q4HCN3 A0A1S3JU34 A0A182WDY7 A0A3B4TYI6 A0A2B4SZC0 A0A2R2MQG4 A0A2R2MQG3 A0A2R2MQF8 A0A182J9S2 A0A0Q3X8Z7 A0A3Q2Z8G1 A0A3Q2XSK6 F1Q6W8 W5L0U3 A0A3Q3J8C4 A0A3B4DAQ5 A0A336LLY4 A0A3N0YML4 A0A336M1K3 H3D688 Q08C18 A0A3Q3MSC5 A0A3B3CZS6 A0A0K8T3A0 A0A1U8DDU9 T1IN54 A0A1U8D6D0 A0A1U7R903 A0A2G8JCI2 A0A3Q2E4V6 A0A182K4S1 F6XU90 G3PWM5 A0A023F648
A0A1J1IQ23 D2A192 U4U670 N6TT07 A0A2J7QS43 A0A067QWM4 Q7PZG7 A0A182SBT9 A0A2J7QS73 A0A182QGI9 A0A182RS06 A0A182XJT9 A0A182UGI4 A0A182L529 A0A182VHN7 A0A182MRP1 A0A182G9L2 A0A182HHJ3 A0A3B3SW41 A0A182HB65 Q17MR4 A0A1S3RSF0 C0H9X4 V9KW78 A0A1W4ZDB2 A0A084WIN3 A0A1B0CA60 H2ZZ55 A0A2G8LLF3 A0A182N2V8 A0A3P8UZ40 V9KJA7 W4XPH5 A0A182FXJ9 A0A3P8X9T0 A0A3B4YF00 A0A3Q1HLH7 A0A3Q1FZW5 A0A3Q1IXQ6 A0A3Q3FLC8 A0A182PP16 W5M531 A0A3P8TRR5 A0A182Y9F8 A0A3Q2VZS3 A0A2D0RI98 E0VDR8 A0A2U9C684 I3IYL8 E1BXI4 A0A3Q1AVS2 E9QDJ5 A0A3P9B089 A0A3B4ZP69 A0A3M6TJD7 A0A1S3JTG6 A0A3Q4HCN3 A0A1S3JU34 A0A182WDY7 A0A3B4TYI6 A0A2B4SZC0 A0A2R2MQG4 A0A2R2MQG3 A0A2R2MQF8 A0A182J9S2 A0A0Q3X8Z7 A0A3Q2Z8G1 A0A3Q2XSK6 F1Q6W8 W5L0U3 A0A3Q3J8C4 A0A3B4DAQ5 A0A336LLY4 A0A3N0YML4 A0A336M1K3 H3D688 Q08C18 A0A3Q3MSC5 A0A3B3CZS6 A0A0K8T3A0 A0A1U8DDU9 T1IN54 A0A1U8D6D0 A0A1U7R903 A0A2G8JCI2 A0A3Q2E4V6 A0A182K4S1 F6XU90 G3PWM5 A0A023F648
Ontologies
KEGG
GO
GO:0016020
GO:0043005
GO:0005930
GO:0036064
GO:0008104
GO:0060271
GO:0046907
GO:0034464
GO:0005737
GO:0015031
GO:1905515
GO:0032402
GO:0007369
GO:0051877
GO:0070121
GO:0001947
GO:0033339
GO:0030030
GO:0016021
GO:0005623
GO:0032436
GO:0005813
GO:0060173
GO:0001750
GO:0001103
GO:0006357
GO:0007224
GO:1903929
GO:0045444
GO:0001654
GO:0007507
GO:0007420
GO:0005634
GO:0007368
GO:0048546
GO:0005515
GO:0005488
GO:0031145
GO:0005506
GO:0030145
GO:0050906
GO:0016818
GO:0003707
GO:0051537
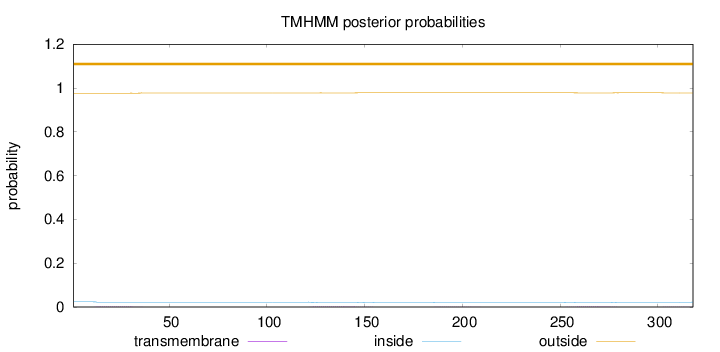
Topology
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13518
Exp number, first 60 AAs:
0.03636
Total prob of N-in:
0.02376
outside
1 - 318
Population Genetic Test Statistics
Pi
164.135714
Theta
159.568836
Tajima's D
0.055022
CLR
14.363177
CSRT
0.390130493475326
Interpretation
Uncertain
