Pre Gene Modal
BGIBMGA001197
Annotation
PREDICTED:_translation_initiation_factor_eIF-2B_subunit_epsilon_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.154
Sequence
CDS
ATGGATAAAGAAAATATCGTACAAGCTGTTGTTATTATGGACACTTTTAACAGTAATTTTTCTCCATTAACAGACAATAAACCAATGGGTTTTCTTGAAGTTGCAGGCGTGCCTCTAATAGATTATGTTCTAGAATCTCTTGCTTTGGGCGGAGTGGGAGAGGCAATTCTGTTTTGTTGTCAAAATGGACAAAAAATAAAAGAACATGTACAGAAACATCAAGATAACAAGTCTCTGTGGAGTCTTACTATGGATATCCAGATTTTGATGTCAGATACTTGTCAGACCATGGGTGATGTGATGAGAGAGTTAGATGCTGCTGCTTTGCTGAAAGGATATTTTGTGCTAGCTGGTATCAATAGCATTACTAATATGAATTTTGCAAGTCTTCTGGAACAACATAAACAAACATGTAAAAAAGACAAAGGGACAGCGATGACTTTGGTTTATAAGAAACTTTCTTGGGAACACCCATTGATAAGCTCAGACAGGTCAACTTTTTTGGCCGCTAACGAGAATACAAAGAAAGTGTTAGTGCACAAGAGATATAGACCTAAGTCCAAGGAGAAGATAATATCTCTTCCTTTGGATTTGGTATTAAATAATTCTGAAGTTAAATTACATCACAATCTGGTTGATACTAACATTGCACTCTGCTCACCAACAGTCCCACCATTGTTCTCGGATAATTTTGATTTTCAAACTAGGGATGACTTCATTCATGGCATATTAATTAATGAGGAAATTCTTGCCAGCTCATTATATTATACATTATTGAAATCAAATCAATATGCTGCTGCTATAACAAATTGGAAAACTTATCAAACAGTTTGTAGAGACATATTACATAAGTGGGTGTATCCACTGTCAATTGAATCGGGTTTGTATTCCCGTGACAAGTATACTCTTACGGGAAATCTTAATTTTGTTGAAAACTCAGCTATGTTGAGCAGATCATGTGTATTAACCGAAAATACATTAATTGGAGCGAAAACGCAAGTACAAGACAACACCACGGTGACAAAATCAATTATCGGAAAGAAATGTACAATTGGAAATAATGTCATAATCAATGGATCACACATAATGAATAATGTAATAATAAAGGACAACTGTAGAATTTCAAACAGTTTTATTGATGACAATTGTGTTATTAGTGAAGGATCAAATCTACATGCTGGCACAATAATTGCAGAAAATGTCAAAGTGGAATCAGGAAGTGATCTCAAAGGCAGAATAGAATGTTCTATTGACAATGATAAAAAACTACAAAAATCAGAATCAAGTGGAACAGAATGGGAAGAGTCATCTGGGAGTAGTGATGAGGAGTTTATTGGTTTTGATAAAGTATGGAGTGACAGTGAATCTTGTTATTCTTCAATCAGTTCAGCAGATTCTTCATTACCTGACTCTCCTGTACCTGATGATACTAAAATGTTCTTGCAAGAAGTCATAGACAGCTTAGCAAGAGGCTATGATGAAAAATTGAAATGCGATTACCTTATATTAGAAATAAATTCATCAAGATATGCCTACAACATTCAGTTAAATGAAGTCAATTTTTTCGTTGTCCGAGCATTACTTAGTATGCCTGTTTTGTCTGAAGCTAAGAATGTGCTCACAACTGTAAAAGATATATTGAAATATTTCCGGCCTGTATTAGCAAACTACATCAAGTCAAAATCTTCTATAATGGATTGCTTACGAGCTGTTGAGGACACCTGTTTAAATTGTGAATGGCTAGATGGAAAATCTGGACAAATTTTACACTTATTATATGAAGCTGATGTAGTAGATGAAGATTCTTTGGTGGATTGGTATGCTGAATTAAAGGAAAATGCAAATCCTTTTGTCAAACAGCCTTCACTTGTAAAATTTTTTGAATGGCTTCAAGAAGCAAGTGAAGAAAGTGATGATTCTGAATAA
Protein
MDKENIVQAVVIMDTFNSNFSPLTDNKPMGFLEVAGVPLIDYVLESLALGGVGEAILFCCQNGQKIKEHVQKHQDNKSLWSLTMDIQILMSDTCQTMGDVMRELDAAALLKGYFVLAGINSITNMNFASLLEQHKQTCKKDKGTAMTLVYKKLSWEHPLISSDRSTFLAANENTKKVLVHKRYRPKSKEKIISLPLDLVLNNSEVKLHHNLVDTNIALCSPTVPPLFSDNFDFQTRDDFIHGILINEEILASSLYYTLLKSNQYAAAITNWKTYQTVCRDILHKWVYPLSIESGLYSRDKYTLTGNLNFVENSAMLSRSCVLTENTLIGAKTQVQDNTTVTKSIIGKKCTIGNNVIINGSHIMNNVIIKDNCRISNSFIDDNCVISEGSNLHAGTIIAENVKVESGSDLKGRIECSIDNDKKLQKSESSGTEWEESSGSSDEEFIGFDKVWSDSESCYSSISSADSSLPDSPVPDDTKMFLQEVIDSLARGYDEKLKCDYLILEINSSRYAYNIQLNEVNFFVVRALLSMPVLSEAKNVLTTVKDILKYFRPVLANYIKSKSSIMDCLRAVEDTCLNCEWLDGKSGQILHLLYEADVVDEDSLVDWYAELKENANPFVKQPSLVKFFEWLQEASEESDDSE
Summary
Uniprot
A0A3S2NQK7
A0A2H1X3L9
S4PY36
A0A212F4G7
A0A194RLS5
A0A194QAF2
+ More
A0A182GFR0 A0A182GWM4 A0A2J7QH87 A0A2R7VZV2 W8AZT4 A0A1Y1NJA6 D2A208 A0A2H8TWI6 A0A131XUG5 J9JNJ7 E9ID55 E1ZZ86 A0A195EP16 A0A195FMG8 A0A195BV38 A0A158NEH5 A0A2A3EI72 A0A154P5S8 A0A151XJ55 A0A2J7QH89 A0A023GD56 E2BLR8 A0A2B4SSI0 A0A3M6U553 A0A0K8W8X4
A0A182GFR0 A0A182GWM4 A0A2J7QH87 A0A2R7VZV2 W8AZT4 A0A1Y1NJA6 D2A208 A0A2H8TWI6 A0A131XUG5 J9JNJ7 E9ID55 E1ZZ86 A0A195EP16 A0A195FMG8 A0A195BV38 A0A158NEH5 A0A2A3EI72 A0A154P5S8 A0A151XJ55 A0A2J7QH89 A0A023GD56 E2BLR8 A0A2B4SSI0 A0A3M6U553 A0A0K8W8X4
Pubmed
EMBL
RSAL01000144
RVE45980.1
ODYU01013242
SOQ59940.1
GAIX01003973
JAA88587.1
+ More
AGBW02010374 OWR48620.1 KQ460205 KPJ16936.1 KQ459299 KPJ01975.1 JXUM01062662 KQ562210 KXJ76401.1 JXUM01093640 KQ564070 KXJ72840.1 NEVH01013984 PNF27948.1 KK854182 PTY12718.1 GAMC01016182 JAB90373.1 GEZM01001289 JAV97984.1 KQ971338 EFA02717.1 GFXV01005723 MBW17528.1 GEFM01006500 JAP69296.1 ABLF02039910 GL762454 EFZ21344.1 GL435311 EFN73501.1 KQ978625 KYN29642.1 KQ981490 KYN41144.1 KQ976401 KYM92447.1 ADTU01013343 KZ288254 PBC30876.1 KQ434822 KZC07203.1 KQ982080 KYQ60371.1 PNF27946.1 GBBM01003614 JAC31804.1 GL449036 EFN83402.1 LSMT01000027 PFX32059.1 RCHS01002248 RMX48578.1 GDHF01005029 JAI47285.1
AGBW02010374 OWR48620.1 KQ460205 KPJ16936.1 KQ459299 KPJ01975.1 JXUM01062662 KQ562210 KXJ76401.1 JXUM01093640 KQ564070 KXJ72840.1 NEVH01013984 PNF27948.1 KK854182 PTY12718.1 GAMC01016182 JAB90373.1 GEZM01001289 JAV97984.1 KQ971338 EFA02717.1 GFXV01005723 MBW17528.1 GEFM01006500 JAP69296.1 ABLF02039910 GL762454 EFZ21344.1 GL435311 EFN73501.1 KQ978625 KYN29642.1 KQ981490 KYN41144.1 KQ976401 KYM92447.1 ADTU01013343 KZ288254 PBC30876.1 KQ434822 KZC07203.1 KQ982080 KYQ60371.1 PNF27946.1 GBBM01003614 JAC31804.1 GL449036 EFN83402.1 LSMT01000027 PFX32059.1 RCHS01002248 RMX48578.1 GDHF01005029 JAI47285.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NQK7
A0A2H1X3L9
S4PY36
A0A212F4G7
A0A194RLS5
A0A194QAF2
+ More
A0A182GFR0 A0A182GWM4 A0A2J7QH87 A0A2R7VZV2 W8AZT4 A0A1Y1NJA6 D2A208 A0A2H8TWI6 A0A131XUG5 J9JNJ7 E9ID55 E1ZZ86 A0A195EP16 A0A195FMG8 A0A195BV38 A0A158NEH5 A0A2A3EI72 A0A154P5S8 A0A151XJ55 A0A2J7QH89 A0A023GD56 E2BLR8 A0A2B4SSI0 A0A3M6U553 A0A0K8W8X4
A0A182GFR0 A0A182GWM4 A0A2J7QH87 A0A2R7VZV2 W8AZT4 A0A1Y1NJA6 D2A208 A0A2H8TWI6 A0A131XUG5 J9JNJ7 E9ID55 E1ZZ86 A0A195EP16 A0A195FMG8 A0A195BV38 A0A158NEH5 A0A2A3EI72 A0A154P5S8 A0A151XJ55 A0A2J7QH89 A0A023GD56 E2BLR8 A0A2B4SSI0 A0A3M6U553 A0A0K8W8X4
PDB
6JLZ
E-value=2.86414e-64,
Score=624
Ontologies
GO
Topology
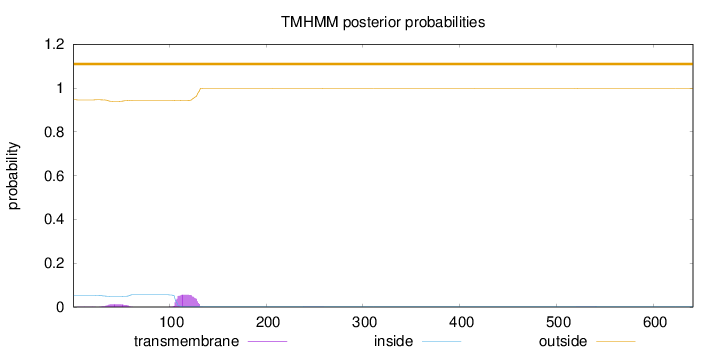
Length:
641
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.48441
Exp number, first 60 AAs:
0.28685
Total prob of N-in:
0.05399
outside
1 - 641
Population Genetic Test Statistics
Pi
198.495322
Theta
178.670971
Tajima's D
0.577493
CLR
0.18389
CSRT
0.539773011349433
Interpretation
Uncertain
