Gene
KWMTBOMO07858
Pre Gene Modal
BGIBMGA001205
Annotation
PREDICTED:_ankyrin_repeat?_SAM_and_basic_leucine_zipper_domain-containing_protein_1_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 2.824
Sequence
CDS
ATGGCTCAATTCAGACCAGCCGGATTTTCCGATGAGGATTCAGATAGTGATGATTACGGATTTTATGAGAAGCCGGTTAGAAACTATGACTATTCTAGTCAAATTAGCAGTAAGAAGAAGGTAGAAGTGGCTTTACAGGAAGCTATCATAAACGGAAACATGAAAGATGTAGAGGATCTTATAAATAATGAATTGAAGAACAATGCTAACATAAAATTGGATAGTGGATGGACCCCACTAATGCATGCATGTTTCCATGCTCAAGATAAAATTGTTCAATATTTATTGGAGAAAGATGCAGATCCTAATTTACACTCAGATTCAGTTACACCTATAATGGCTGCATGCTCAAATAGTAGTGCAGATAATGACACAATATACAGTATTGTGTGTAACCTCATAGACAGAAATTGCATACTGAATATTGGAGATAGATATGGACAAACCCCACTAATGAGGGCTATAAGCAGCGGCCGTGTGGCAGTGGCTCAAAAATTACTAGATATGCAAGTTAATATAGAAATGAGGGATCAGCATGGGTGGACAGCTGTTTTCTGGGCTGTCCACCACAATCAACCAGAAATACTAGAAATGTTAATTGAAAAAGGAGCTAGACTAACTGAAGTTGACAAATCGAACCGAACGCTATTAGAAATTGCCAATTGTCATGATCATCAAGTGATAATTGAAATATTAAAGAAATCACTCAACCTAGACGATGATCATAAAACAGATGAAAACTCATACTTAAACCATCAATTAACATCTTGGCAAGATTATTATCCGGGAATTGAGAAAAATGAACGACCGAACTATAAAAACGAAATTCAGAATCTGCTTTACGGCATGAGCTGTGAACGTTTGACGCCCCTGATCGTAACGTCCGGCATCGATTTGAGGACGTTCCTACTGCTGGACGCCGAGGAAATGGTTAAATTAGGTATAGAGATGCCATACGAAAGGCACAGACTGAAATACGGATTACGGAATTTCCATTTGAGAGGATGGAAATTAAACGCGGTGGCCGGTTTGTATGCTCGGAAAATGGATAACTATTGCGTTTTAGACTGTTTAACTTCATTGGGGAGTCATTTACATCAGATTTACATACTAGAGGCCACGCTACAATATATTCTACGCGAGTACGCTAAAGTCGAAAATCAAATAAAACACCAACCTCCGGACTCGCCTTTACTTAAAAAATTGAAAACATCAACAAAGAAAATGCTCACTAACATAAACAGCATACGAAGGGAAATTAAAAGCATGAAAATTACTTTACAGAAGATTGATAGTAATAATCCTCGCCCCGCCGATCTGCTACCAAAGAAGTCAACTCAAGAAATTGCTATCCAATATATCACAAAAGCCGTCTCGCTTTGCTCTCTAGCCTTTATACTATATCAAGGACGAACCTTGTTGGGCAATTTGTTAAAAAAATAA
Protein
MAQFRPAGFSDEDSDSDDYGFYEKPVRNYDYSSQISSKKKVEVALQEAIINGNMKDVEDLINNELKNNANIKLDSGWTPLMHACFHAQDKIVQYLLEKDADPNLHSDSVTPIMAACSNSSADNDTIYSIVCNLIDRNCILNIGDRYGQTPLMRAISSGRVAVAQKLLDMQVNIEMRDQHGWTAVFWAVHHNQPEILEMLIEKGARLTEVDKSNRTLLEIANCHDHQVIIEILKKSLNLDDDHKTDENSYLNHQLTSWQDYYPGIEKNERPNYKNEIQNLLYGMSCERLTPLIVTSGIDLRTFLLLDAEEMVKLGIEMPYERHRLKYGLRNFHLRGWKLNAVAGLYARKMDNYCVLDCLTSLGSHLHQIYILEATLQYILREYAKVENQIKHQPPDSPLLKKLKTSTKKMLTNINSIRREIKSMKITLQKIDSNNPRPADLLPKKSTQEIAIQYITKAVSLCSLAFILYQGRTLLGNLLKK
Summary
Uniprot
H9IVC5
A0A194RLR6
A0A194QEU4
A0A2A4JKL9
A0A2A4JM82
A0A2H1VHK0
+ More
A0A2H1V2H2 A0A3S2NAN9 A0A212FNF1 A0A1Q3F3M7 A0A0J9R7V9 B4HRL2 A0A1B0GJA4 A0A182GXJ6 B0X6F6 W8CB08 A0A1I8Q181 A0A023ESK7 A0A1B0BSZ7 A0A1W4WPV2 A0A1A9ZYJ3 A0A1A9V9R0 A0A0K8UI97 B4MRE5 D6WAG4 A0A195B091 A0A1I8MP99 Q7K5K0 A0A151IJS3 A0A0T6B1E7 A0A034WVC6 A0A158NKJ4 A0A0P4YWU0 B3MGU3 B4P2P6 A0A336LL94 A0A1S4EVF5 Q17PP6 A0A1L8DSL6 A0A1A9W6X2 A0A0A1X3V6 A0A0P4YU91 A0A1A9XP45 E9G070 A0A0P5BFN3 A0A164U921 W8BWI1 A0A0P6IMU2 A0A1J1IZZ3 A0A0M8ZUF2 A0A1Y1KSB5 A0A182RJT8 A0A088A2H5 A0A151JB38 A0A0P5NVU3 A0A182YIU6 A0A182W3Y0 A0A182IEQ7 Q7QDY6 A0A1B0G4H4 A0A195FC58 A0A0M4EFD8 R4WDL9 A0A2A3E9Q7 A0A182J2G2 A0A084VUQ9 B4LL74 A0A1W4WBN0 B3N8T1 B4J5M1 A0A0N8C3A8 K7JL29 A0A182KZZ5 A0A2J7QNZ5 T1HCD1 A0A154NW71 A0A0P5IMA3 A0A210R3U4 A0A2T7NT70 B4KPH0 E9IFC8 A0A1B6FAW1
A0A2H1V2H2 A0A3S2NAN9 A0A212FNF1 A0A1Q3F3M7 A0A0J9R7V9 B4HRL2 A0A1B0GJA4 A0A182GXJ6 B0X6F6 W8CB08 A0A1I8Q181 A0A023ESK7 A0A1B0BSZ7 A0A1W4WPV2 A0A1A9ZYJ3 A0A1A9V9R0 A0A0K8UI97 B4MRE5 D6WAG4 A0A195B091 A0A1I8MP99 Q7K5K0 A0A151IJS3 A0A0T6B1E7 A0A034WVC6 A0A158NKJ4 A0A0P4YWU0 B3MGU3 B4P2P6 A0A336LL94 A0A1S4EVF5 Q17PP6 A0A1L8DSL6 A0A1A9W6X2 A0A0A1X3V6 A0A0P4YU91 A0A1A9XP45 E9G070 A0A0P5BFN3 A0A164U921 W8BWI1 A0A0P6IMU2 A0A1J1IZZ3 A0A0M8ZUF2 A0A1Y1KSB5 A0A182RJT8 A0A088A2H5 A0A151JB38 A0A0P5NVU3 A0A182YIU6 A0A182W3Y0 A0A182IEQ7 Q7QDY6 A0A1B0G4H4 A0A195FC58 A0A0M4EFD8 R4WDL9 A0A2A3E9Q7 A0A182J2G2 A0A084VUQ9 B4LL74 A0A1W4WBN0 B3N8T1 B4J5M1 A0A0N8C3A8 K7JL29 A0A182KZZ5 A0A2J7QNZ5 T1HCD1 A0A154NW71 A0A0P5IMA3 A0A210R3U4 A0A2T7NT70 B4KPH0 E9IFC8 A0A1B6FAW1
Pubmed
19121390
26354079
22118469
22936249
17994087
26483478
+ More
24495485 24945155 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 21347285 17550304 17510324 25830018 21292972 28004739 25244985 12364791 14747013 17210077 23691247 24438588 20075255 20966253 28812685 21282665
24495485 24945155 18362917 19820115 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 21347285 17550304 17510324 25830018 21292972 28004739 25244985 12364791 14747013 17210077 23691247 24438588 20075255 20966253 28812685 21282665
EMBL
BABH01000868
BABH01000869
KQ460205
KPJ16926.1
KQ459299
KPJ01986.1
+ More
NWSH01001121 PCG72501.1 PCG72502.1 ODYU01002593 SOQ40297.1 ODYU01000370 SOQ35040.1 RSAL01000144 RVE45989.1 AGBW02005699 OWR55265.1 GFDL01012880 JAV22165.1 CM002911 KMY92222.1 CH480816 EDW46894.1 AJWK01019999 JXUM01020581 KQ560576 KXJ81835.1 DS232412 EDS41439.1 GAMC01000874 JAC05682.1 GAPW01001333 JAC12265.1 JXJN01019947 GDHF01033303 GDHF01026058 JAI19011.1 JAI26256.1 CH963850 EDW74684.1 KQ971312 EEZ98003.2 KQ976692 KYM77876.1 AE013599 AY047578 AAF59118.2 AAK77310.1 KQ977294 KYN03899.1 LJIG01016338 KRT80901.1 GAKP01000792 JAC58160.1 ADTU01018869 GDIP01226024 JAI97377.1 CH902619 EDV37861.1 CM000157 EDW89307.1 UFQT01000039 SSX18475.1 CH477190 EAT48705.1 GFDF01004616 JAV09468.1 GBXI01008263 JAD06029.1 GDIP01223522 JAI99879.1 GL732528 EFX87449.1 GDIP01185392 JAJ38010.1 LRGB01001581 KZS11155.1 GAMC01000875 JAC05681.1 GDIQ01008616 JAN86121.1 CVRI01000065 CRL05835.1 KQ435881 KOX69823.1 GEZM01076998 JAV63478.1 KQ979182 KYN22326.1 GDIQ01137028 JAL14698.1 APCN01009333 AAAB01008849 EAA07102.4 CCAG010007755 KQ981693 KYN37787.1 CP012524 ALC40897.1 AK417808 BAN21023.1 KZ288311 PBC28458.1 ATLV01016988 KE525139 KFB41703.1 CH940648 EDW60811.1 CH954177 EDV59558.1 CH916367 EDW00784.1 GDIQ01118995 JAL32731.1 NEVH01012098 PNF30302.1 ACPB03002116 KQ434772 KZC03916.1 GDIQ01212015 JAK39710.1 NEDP02000506 OWF55730.1 PZQS01000009 PVD24361.1 CH933808 EDW10166.1 GL762838 EFZ20703.1 GECZ01022447 JAS47322.1
NWSH01001121 PCG72501.1 PCG72502.1 ODYU01002593 SOQ40297.1 ODYU01000370 SOQ35040.1 RSAL01000144 RVE45989.1 AGBW02005699 OWR55265.1 GFDL01012880 JAV22165.1 CM002911 KMY92222.1 CH480816 EDW46894.1 AJWK01019999 JXUM01020581 KQ560576 KXJ81835.1 DS232412 EDS41439.1 GAMC01000874 JAC05682.1 GAPW01001333 JAC12265.1 JXJN01019947 GDHF01033303 GDHF01026058 JAI19011.1 JAI26256.1 CH963850 EDW74684.1 KQ971312 EEZ98003.2 KQ976692 KYM77876.1 AE013599 AY047578 AAF59118.2 AAK77310.1 KQ977294 KYN03899.1 LJIG01016338 KRT80901.1 GAKP01000792 JAC58160.1 ADTU01018869 GDIP01226024 JAI97377.1 CH902619 EDV37861.1 CM000157 EDW89307.1 UFQT01000039 SSX18475.1 CH477190 EAT48705.1 GFDF01004616 JAV09468.1 GBXI01008263 JAD06029.1 GDIP01223522 JAI99879.1 GL732528 EFX87449.1 GDIP01185392 JAJ38010.1 LRGB01001581 KZS11155.1 GAMC01000875 JAC05681.1 GDIQ01008616 JAN86121.1 CVRI01000065 CRL05835.1 KQ435881 KOX69823.1 GEZM01076998 JAV63478.1 KQ979182 KYN22326.1 GDIQ01137028 JAL14698.1 APCN01009333 AAAB01008849 EAA07102.4 CCAG010007755 KQ981693 KYN37787.1 CP012524 ALC40897.1 AK417808 BAN21023.1 KZ288311 PBC28458.1 ATLV01016988 KE525139 KFB41703.1 CH940648 EDW60811.1 CH954177 EDV59558.1 CH916367 EDW00784.1 GDIQ01118995 JAL32731.1 NEVH01012098 PNF30302.1 ACPB03002116 KQ434772 KZC03916.1 GDIQ01212015 JAK39710.1 NEDP02000506 OWF55730.1 PZQS01000009 PVD24361.1 CH933808 EDW10166.1 GL762838 EFZ20703.1 GECZ01022447 JAS47322.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000001292 UP000092461 UP000069940 UP000249989 UP000002320 UP000095300 UP000092460 UP000192223 UP000092445 UP000078200 UP000007798 UP000007266 UP000078540 UP000095301 UP000000803 UP000078542 UP000005205 UP000007801 UP000002282 UP000008820 UP000091820 UP000092443 UP000000305 UP000076858 UP000183832 UP000053105 UP000075900 UP000005203 UP000078492 UP000076408 UP000075920 UP000075840 UP000007062 UP000092444 UP000078541 UP000092553 UP000242457 UP000075880 UP000030765 UP000008792 UP000192221 UP000008711 UP000001070 UP000002358 UP000075882 UP000235965 UP000015103 UP000076502 UP000242188 UP000245119 UP000009192
UP000001292 UP000092461 UP000069940 UP000249989 UP000002320 UP000095300 UP000092460 UP000192223 UP000092445 UP000078200 UP000007798 UP000007266 UP000078540 UP000095301 UP000000803 UP000078542 UP000005205 UP000007801 UP000002282 UP000008820 UP000091820 UP000092443 UP000000305 UP000076858 UP000183832 UP000053105 UP000075900 UP000005203 UP000078492 UP000076408 UP000075920 UP000075840 UP000007062 UP000092444 UP000078541 UP000092553 UP000242457 UP000075880 UP000030765 UP000008792 UP000192221 UP000008711 UP000001070 UP000002358 UP000075882 UP000235965 UP000015103 UP000076502 UP000242188 UP000245119 UP000009192
Pfam
Interpro
IPR020683
Ankyrin_rpt-contain_dom
+ More
IPR036770 Ankyrin_rpt-contain_sf
IPR013761 SAM/pointed_sf
IPR002110 Ankyrin_rpt
IPR001660 SAM
IPR002099 DNA_mismatch_repair_N
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR042121 MutL_C_regsub
IPR014790 MutL_C
IPR042120 MutL_C_dimsub
IPR036890 HATPase_C_sf
IPR003594 HATPase_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR014762 DNA_mismatch_repair_CS
IPR038973 MutL/Mlh/Pms
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
IPR036770 Ankyrin_rpt-contain_sf
IPR013761 SAM/pointed_sf
IPR002110 Ankyrin_rpt
IPR001660 SAM
IPR002099 DNA_mismatch_repair_N
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR042121 MutL_C_regsub
IPR014790 MutL_C
IPR042120 MutL_C_dimsub
IPR036890 HATPase_C_sf
IPR003594 HATPase_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR014762 DNA_mismatch_repair_CS
IPR038973 MutL/Mlh/Pms
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
SUPFAM
ProteinModelPortal
H9IVC5
A0A194RLR6
A0A194QEU4
A0A2A4JKL9
A0A2A4JM82
A0A2H1VHK0
+ More
A0A2H1V2H2 A0A3S2NAN9 A0A212FNF1 A0A1Q3F3M7 A0A0J9R7V9 B4HRL2 A0A1B0GJA4 A0A182GXJ6 B0X6F6 W8CB08 A0A1I8Q181 A0A023ESK7 A0A1B0BSZ7 A0A1W4WPV2 A0A1A9ZYJ3 A0A1A9V9R0 A0A0K8UI97 B4MRE5 D6WAG4 A0A195B091 A0A1I8MP99 Q7K5K0 A0A151IJS3 A0A0T6B1E7 A0A034WVC6 A0A158NKJ4 A0A0P4YWU0 B3MGU3 B4P2P6 A0A336LL94 A0A1S4EVF5 Q17PP6 A0A1L8DSL6 A0A1A9W6X2 A0A0A1X3V6 A0A0P4YU91 A0A1A9XP45 E9G070 A0A0P5BFN3 A0A164U921 W8BWI1 A0A0P6IMU2 A0A1J1IZZ3 A0A0M8ZUF2 A0A1Y1KSB5 A0A182RJT8 A0A088A2H5 A0A151JB38 A0A0P5NVU3 A0A182YIU6 A0A182W3Y0 A0A182IEQ7 Q7QDY6 A0A1B0G4H4 A0A195FC58 A0A0M4EFD8 R4WDL9 A0A2A3E9Q7 A0A182J2G2 A0A084VUQ9 B4LL74 A0A1W4WBN0 B3N8T1 B4J5M1 A0A0N8C3A8 K7JL29 A0A182KZZ5 A0A2J7QNZ5 T1HCD1 A0A154NW71 A0A0P5IMA3 A0A210R3U4 A0A2T7NT70 B4KPH0 E9IFC8 A0A1B6FAW1
A0A2H1V2H2 A0A3S2NAN9 A0A212FNF1 A0A1Q3F3M7 A0A0J9R7V9 B4HRL2 A0A1B0GJA4 A0A182GXJ6 B0X6F6 W8CB08 A0A1I8Q181 A0A023ESK7 A0A1B0BSZ7 A0A1W4WPV2 A0A1A9ZYJ3 A0A1A9V9R0 A0A0K8UI97 B4MRE5 D6WAG4 A0A195B091 A0A1I8MP99 Q7K5K0 A0A151IJS3 A0A0T6B1E7 A0A034WVC6 A0A158NKJ4 A0A0P4YWU0 B3MGU3 B4P2P6 A0A336LL94 A0A1S4EVF5 Q17PP6 A0A1L8DSL6 A0A1A9W6X2 A0A0A1X3V6 A0A0P4YU91 A0A1A9XP45 E9G070 A0A0P5BFN3 A0A164U921 W8BWI1 A0A0P6IMU2 A0A1J1IZZ3 A0A0M8ZUF2 A0A1Y1KSB5 A0A182RJT8 A0A088A2H5 A0A151JB38 A0A0P5NVU3 A0A182YIU6 A0A182W3Y0 A0A182IEQ7 Q7QDY6 A0A1B0G4H4 A0A195FC58 A0A0M4EFD8 R4WDL9 A0A2A3E9Q7 A0A182J2G2 A0A084VUQ9 B4LL74 A0A1W4WBN0 B3N8T1 B4J5M1 A0A0N8C3A8 K7JL29 A0A182KZZ5 A0A2J7QNZ5 T1HCD1 A0A154NW71 A0A0P5IMA3 A0A210R3U4 A0A2T7NT70 B4KPH0 E9IFC8 A0A1B6FAW1
PDB
6NE2
E-value=1.47586e-14,
Score=194
Ontologies
GO
PANTHER
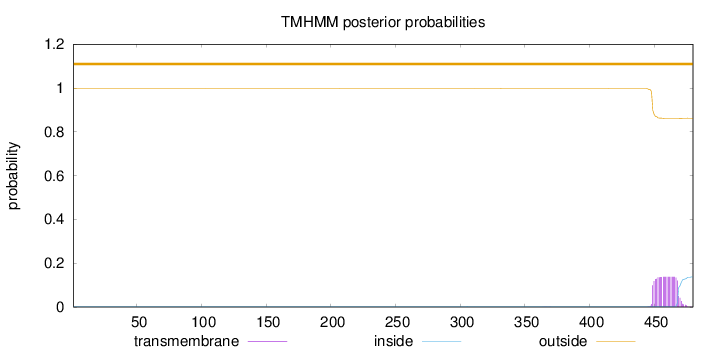
Topology
Length:
480
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.8466
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00262
outside
1 - 480
Population Genetic Test Statistics
Pi
290.404132
Theta
217.893058
Tajima's D
1.437794
CLR
0.293601
CSRT
0.774561271936403
Interpretation
Uncertain
