Gene
KWMTBOMO07855 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001206
Annotation
chaperonin_containing_t-complex_polypeptide_1_beta_subunit_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.581
Sequence
CDS
ATGGTATCATTAAATCCAATTCGTATCCTTAAAAATGAAGCAGAAGAGGAGAAAGCTGAAGTTGCCCGGATGTCCAGTTTCATCGGTGCTATTGCCATAGGAGATTTGGTAAAGAGTACTTTAGGTCCCAAAGGAATGGATAAGATCTTAGTTTCCTGCGGGAGGAACTCTGGTCAGGTCGAAGTAACAAATGATGGTGCTACGATATTGAAATCTATCGGTGTGGATAATCCTGCTGCTAAAATCCTAGTAGATATGTCTAAGGTACAAGATGAAGAAGTCGGAGATGGCACCACATCTGTTACAGTTTTGGCTGCTGAATTACTTCGAGAGGCTGAAAAATTAATTGAACAAAAACTGCACCCTCAAACAATCATTGCTGGTTGGAGAATTGCTTCTGATGCAGCCAAACAAGCTCTTGCAGAAGCTAGCTTTGATCATCAGAAGAACTTAAATGAGGCTTCATTAAGAGTGGATTTAGAAAACATCGCACGGACCACACTGAGCTCAAAGATCCTTTCAAATCACAAAGAGCATTTCACAAAATTAGCAGTTGATGCAGTTTTGCGTTTGAAAGGATCTGGTAATTTAAAAGCTATCCAAATTATCAAAATATCTGGAGGTCTTCTTGAAGAGTCATTCTTGGATGAAGGGTTCTTGCTAAACAAAAAAGTTGGTGTACATCAACCAAAGAAAGTAGAAAATGCTAACATCCTTATTGCAAACACACCAATGGACACTGACAAGATTAAAGTGTTTGGTTCCACAATTAAAGTGGATTCAATGGCCAAGATTGCTGAATTGGAAGTAGCTGAGAAAGAGAAAATGAAGGACAAAGTGAACAAGATTTTAGCACACAAATGTAATGTCTTCATTAACAGACAACTTATATACAACTACCCAGAGCAGTTGTTTGCTGATGCTGGAGTAATGGCAATTGAGCATGCTGATTTCGAAGGTATCGAAAGGCTTGGCTTGGTCACTGGAGGGGAAATTGTGTCAACATTTGACTCACCAGACAAAGTGAAACTTGGTCACTGCAAATTGATCGAAGAGGTTCTGATTGGTGATGAGAGCTTGATCCGCTTCTCTGGTGTAGCGTTGGGCTCGGCTTGCACGATCGTAATCCGTGGTGCCACGCAACAAGTTATCGACGAAGCTGAGCGTTCACTCCATGATGCGCTTTGCGTGCTTGCTGCTACTGTAAAAGAACCGAAGGTCATATGTGGAGGAGGGGCTGGTGAAATGCTAATGGCAGAAGCAGCTTCCCGCGCCGGCGCTCGCACAGCCGGGAAGGAAGCCGCAGCAGCCGAAGCCTTTGCAGTCGCGCTCCGGAGACTGCCGTCCGCTGTCGCCGACAATGCTGGCTACGACAGTGCTGATCTCATTGCTCGACTCAGAGCTCATCATTCTCGAGGAGAAAACACTATGGGATTAGATATGCAAAACGGCACTGTTGGTGATATGAAGAAGTTGGGTGTGACCGAGTCGTATGTCGTAAAGAGACAGGTCCTGTTGTCTGCGGCTGAAGCGGCCGAGATGATTCTTCGTGTCGATAACATTCTGAAGGCAGCACCCAGACGCCGCGGACCCGACCGTCGCCCATGTTAA
Protein
MVSLNPIRILKNEAEEEKAEVARMSSFIGAIAIGDLVKSTLGPKGMDKILVSCGRNSGQVEVTNDGATILKSIGVDNPAAKILVDMSKVQDEEVGDGTTSVTVLAAELLREAEKLIEQKLHPQTIIAGWRIASDAAKQALAEASFDHQKNLNEASLRVDLENIARTTLSSKILSNHKEHFTKLAVDAVLRLKGSGNLKAIQIIKISGGLLEESFLDEGFLLNKKVGVHQPKKVENANILIANTPMDTDKIKVFGSTIKVDSMAKIAELEVAEKEKMKDKVNKILAHKCNVFINRQLIYNYPEQLFADAGVMAIEHADFEGIERLGLVTGGEIVSTFDSPDKVKLGHCKLIEEVLIGDESLIRFSGVALGSACTIVIRGATQQVIDEAERSLHDALCVLAATVKEPKVICGGGAGEMLMAEAASRAGARTAGKEAAAAEAFAVALRRLPSAVADNAGYDSADLIARLRAHHSRGENTMGLDMQNGTVGDMKKLGVTESYVVKRQVLLSAAEAAEMILRVDNILKAAPRRRGPDRRPC
Summary
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
Q2F6C2
A0A2H1V2G3
A0A2A4JL50
S4PDE5
A0A212ESF7
A0A194RGI0
+ More
A0A194Q8Z1 A0A182UXK1 A0A182TK59 A0A182LMA0 A0A182HI69 A0A1B6CS56 A0A1B6GB87 Q7Q1P2 A0A182QGJ2 D6WBW7 U5EZ38 A0A182YFT8 A0A182KFJ9 A0A1B6LBH7 A0A023ETP2 A0A1S4FHA0 V5GNR1 A0A1Y1L6K5 A0A182M9I4 A0A182VU05 A0A182RVZ2 A0A182XN24 A0A182PTU9 A0A2J7RCP8 A0A1L8DUA4 V5RF49 A0A1W4X083 A0A336M8P4 A0A336MS14 T1PFF7 A0A1Q3FWH6 A0A182NBG3 T1E7V0 A0A182F260 A0A2M4BJV3 W5JEP1 E0VTG8 A0A0A1WR41 A0A1B0CV76 D3TMR1 A0A0L0CJW2 A0A034WLE7 A0A1J1IQ64 A0A088AAV5 F2D638 A0A0K8WEE3 A0A0K8U9A7 A0A1L8EDC8 A0A1I8P7C9 A0A1A9UEE5 A0A2H8U143 A0A2S2NU54 A0A0N0BHT9 A0A1L8EDQ5 A0A0K8TSS4 K7J2G8 J9K0Z9 A0A2A3EE83 W8BBM5 A0A1B0FGM9 A0A310SE98 A0A2M4A9Q8 A0A067RHG6 A0A2H8TFP5 A0A1B0DIB3 A0A1B0ADK9 F4WS67 A0A0M5JAW2 A0A158N9W9 A0A0L7QTR4 A0A1A9YS69 A0A151WRU9 A0A151ISP9 A0A182J824 A0A1A9X0B7 J9JK95 A0A0C9QE42 A0A0C9RYP3 J3JUX5 A0A1Q3FX16 N6U469 A0A3B0KWY2 B4M281 B4JIV9 A0A3L8E321 B3MZK0 A0A1W4UD59 E2BQ93 E2AE48 Q29GX0 B4IJQ6 B3NUH9 B4PZY2 B4N1Z4
A0A194Q8Z1 A0A182UXK1 A0A182TK59 A0A182LMA0 A0A182HI69 A0A1B6CS56 A0A1B6GB87 Q7Q1P2 A0A182QGJ2 D6WBW7 U5EZ38 A0A182YFT8 A0A182KFJ9 A0A1B6LBH7 A0A023ETP2 A0A1S4FHA0 V5GNR1 A0A1Y1L6K5 A0A182M9I4 A0A182VU05 A0A182RVZ2 A0A182XN24 A0A182PTU9 A0A2J7RCP8 A0A1L8DUA4 V5RF49 A0A1W4X083 A0A336M8P4 A0A336MS14 T1PFF7 A0A1Q3FWH6 A0A182NBG3 T1E7V0 A0A182F260 A0A2M4BJV3 W5JEP1 E0VTG8 A0A0A1WR41 A0A1B0CV76 D3TMR1 A0A0L0CJW2 A0A034WLE7 A0A1J1IQ64 A0A088AAV5 F2D638 A0A0K8WEE3 A0A0K8U9A7 A0A1L8EDC8 A0A1I8P7C9 A0A1A9UEE5 A0A2H8U143 A0A2S2NU54 A0A0N0BHT9 A0A1L8EDQ5 A0A0K8TSS4 K7J2G8 J9K0Z9 A0A2A3EE83 W8BBM5 A0A1B0FGM9 A0A310SE98 A0A2M4A9Q8 A0A067RHG6 A0A2H8TFP5 A0A1B0DIB3 A0A1B0ADK9 F4WS67 A0A0M5JAW2 A0A158N9W9 A0A0L7QTR4 A0A1A9YS69 A0A151WRU9 A0A151ISP9 A0A182J824 A0A1A9X0B7 J9JK95 A0A0C9QE42 A0A0C9RYP3 J3JUX5 A0A1Q3FX16 N6U469 A0A3B0KWY2 B4M281 B4JIV9 A0A3L8E321 B3MZK0 A0A1W4UD59 E2BQ93 E2AE48 Q29GX0 B4IJQ6 B3NUH9 B4PZY2 B4N1Z4
Pubmed
19121390
23622113
22118469
26354079
20966253
12364791
+ More
18362917 19820115 25244985 24945155 26483478 28004739 25315136 20920257 23761445 20566863 25830018 20353571 26108605 25348373 21415278 26369729 20075255 24495485 24845553 21719571 21347285 22516182 23537049 17994087 30249741 20798317 15632085 17550304
18362917 19820115 25244985 24945155 26483478 28004739 25315136 20920257 23761445 20566863 25830018 20353571 26108605 25348373 21415278 26369729 20075255 24495485 24845553 21719571 21347285 22516182 23537049 17994087 30249741 20798317 15632085 17550304
EMBL
BABH01000874
DQ311150
ABD36095.1
ODYU01000369
SOQ35035.1
NWSH01001121
+ More
PCG72496.1 GAIX01004977 JAA87583.1 AGBW02012794 OWR44428.1 KQ460205 KPJ16923.1 KQ459299 KPJ01988.1 APCN01005638 GEDC01021040 JAS16258.1 GECZ01010074 JAS59695.1 AAAB01008980 EAA14559.4 AXCN02000619 KQ971315 EEZ97857.1 GANO01001489 JAB58382.1 GEBQ01019093 JAT20884.1 JXUM01135353 GAPW01000888 KQ568258 JAC12710.1 KXJ69077.1 GALX01002697 JAB65769.1 GEZM01068909 JAV66707.1 AXCM01002101 NEVH01005885 PNF38611.1 GFDF01004062 JAV10022.1 KF621133 AHB33469.1 UFQT01000702 SSX26646.1 UFQT01002349 SSX33252.1 KA646643 AFP61272.1 GFDL01003081 JAV31964.1 GAMD01003328 JAA98262.1 GGFJ01004143 MBW53284.1 ADMH02001369 ETN62822.1 DS235765 EEB16674.1 GBXI01013302 JAD00990.1 AJWK01030162 AJWK01030163 EZ422713 ADD18989.1 JRES01000302 KNC32535.1 GAKP01004359 JAC54593.1 CVRI01000057 CRL02381.1 AK359348 BAJ90559.1 GDHF01002776 JAI49538.1 GDHF01029374 JAI22940.1 GFDG01002062 JAV16737.1 GFXV01008104 MBW19909.1 GGMR01007969 MBY20588.1 KQ435742 KOX76688.1 GFDG01002060 JAV16739.1 GDAI01000427 JAI17176.1 ABLF02030785 KZ288267 PBC30093.1 GAMC01016029 GAMC01016027 JAB90526.1 CCAG010019381 KQ770716 OAD52605.1 GGFK01004170 MBW37491.1 KK852506 KDR22488.1 GFXV01001138 MBW12943.1 AJVK01034336 GL888300 EGI62961.1 CP012528 ALC49661.1 ADTU01009804 KQ414742 KOC61944.1 KQ982796 KYQ50586.1 KQ981064 KYN09856.1 ABLF02011588 ABLF02011589 GBYB01012673 JAG82440.1 GBYB01014410 JAG84177.1 BT127040 KB632379 AEE62002.1 ERL94112.1 GFDL01003082 JAV31963.1 APGK01040371 KB740979 ENN76400.1 OUUW01000015 SPP88558.1 CH940651 EDW65785.2 CH916370 EDV99523.1 QOIP01000001 RLU27131.1 CH902635 EDV33801.2 GL449698 EFN82147.1 GL438827 EFN68281.1 CH379064 EAL31989.2 CH480849 EDW51264.1 CH954180 EDV46302.2 CM000162 EDX02188.2 CH963925 EDW78383.2
PCG72496.1 GAIX01004977 JAA87583.1 AGBW02012794 OWR44428.1 KQ460205 KPJ16923.1 KQ459299 KPJ01988.1 APCN01005638 GEDC01021040 JAS16258.1 GECZ01010074 JAS59695.1 AAAB01008980 EAA14559.4 AXCN02000619 KQ971315 EEZ97857.1 GANO01001489 JAB58382.1 GEBQ01019093 JAT20884.1 JXUM01135353 GAPW01000888 KQ568258 JAC12710.1 KXJ69077.1 GALX01002697 JAB65769.1 GEZM01068909 JAV66707.1 AXCM01002101 NEVH01005885 PNF38611.1 GFDF01004062 JAV10022.1 KF621133 AHB33469.1 UFQT01000702 SSX26646.1 UFQT01002349 SSX33252.1 KA646643 AFP61272.1 GFDL01003081 JAV31964.1 GAMD01003328 JAA98262.1 GGFJ01004143 MBW53284.1 ADMH02001369 ETN62822.1 DS235765 EEB16674.1 GBXI01013302 JAD00990.1 AJWK01030162 AJWK01030163 EZ422713 ADD18989.1 JRES01000302 KNC32535.1 GAKP01004359 JAC54593.1 CVRI01000057 CRL02381.1 AK359348 BAJ90559.1 GDHF01002776 JAI49538.1 GDHF01029374 JAI22940.1 GFDG01002062 JAV16737.1 GFXV01008104 MBW19909.1 GGMR01007969 MBY20588.1 KQ435742 KOX76688.1 GFDG01002060 JAV16739.1 GDAI01000427 JAI17176.1 ABLF02030785 KZ288267 PBC30093.1 GAMC01016029 GAMC01016027 JAB90526.1 CCAG010019381 KQ770716 OAD52605.1 GGFK01004170 MBW37491.1 KK852506 KDR22488.1 GFXV01001138 MBW12943.1 AJVK01034336 GL888300 EGI62961.1 CP012528 ALC49661.1 ADTU01009804 KQ414742 KOC61944.1 KQ982796 KYQ50586.1 KQ981064 KYN09856.1 ABLF02011588 ABLF02011589 GBYB01012673 JAG82440.1 GBYB01014410 JAG84177.1 BT127040 KB632379 AEE62002.1 ERL94112.1 GFDL01003082 JAV31963.1 APGK01040371 KB740979 ENN76400.1 OUUW01000015 SPP88558.1 CH940651 EDW65785.2 CH916370 EDV99523.1 QOIP01000001 RLU27131.1 CH902635 EDV33801.2 GL449698 EFN82147.1 GL438827 EFN68281.1 CH379064 EAL31989.2 CH480849 EDW51264.1 CH954180 EDV46302.2 CM000162 EDX02188.2 CH963925 EDW78383.2
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000075903
+ More
UP000075902 UP000075882 UP000075840 UP000007062 UP000075886 UP000007266 UP000076408 UP000075881 UP000069940 UP000249989 UP000075883 UP000075920 UP000075900 UP000076407 UP000075885 UP000235965 UP000192223 UP000095301 UP000075884 UP000069272 UP000000673 UP000009046 UP000092461 UP000037069 UP000183832 UP000005203 UP000095300 UP000078200 UP000053105 UP000002358 UP000007819 UP000242457 UP000092444 UP000027135 UP000092462 UP000092445 UP000007755 UP000092553 UP000005205 UP000053825 UP000092443 UP000075809 UP000078492 UP000075880 UP000091820 UP000030742 UP000019118 UP000268350 UP000008792 UP000001070 UP000279307 UP000007801 UP000192221 UP000008237 UP000000311 UP000001819 UP000001292 UP000008711 UP000002282 UP000007798
UP000075902 UP000075882 UP000075840 UP000007062 UP000075886 UP000007266 UP000076408 UP000075881 UP000069940 UP000249989 UP000075883 UP000075920 UP000075900 UP000076407 UP000075885 UP000235965 UP000192223 UP000095301 UP000075884 UP000069272 UP000000673 UP000009046 UP000092461 UP000037069 UP000183832 UP000005203 UP000095300 UP000078200 UP000053105 UP000002358 UP000007819 UP000242457 UP000092444 UP000027135 UP000092462 UP000092445 UP000007755 UP000092553 UP000005205 UP000053825 UP000092443 UP000075809 UP000078492 UP000075880 UP000091820 UP000030742 UP000019118 UP000268350 UP000008792 UP000001070 UP000279307 UP000007801 UP000192221 UP000008237 UP000000311 UP000001819 UP000001292 UP000008711 UP000002282 UP000007798
PRIDE
Pfam
PF00118 Cpn60_TCP1
Interpro
Gene 3D
ProteinModelPortal
Q2F6C2
A0A2H1V2G3
A0A2A4JL50
S4PDE5
A0A212ESF7
A0A194RGI0
+ More
A0A194Q8Z1 A0A182UXK1 A0A182TK59 A0A182LMA0 A0A182HI69 A0A1B6CS56 A0A1B6GB87 Q7Q1P2 A0A182QGJ2 D6WBW7 U5EZ38 A0A182YFT8 A0A182KFJ9 A0A1B6LBH7 A0A023ETP2 A0A1S4FHA0 V5GNR1 A0A1Y1L6K5 A0A182M9I4 A0A182VU05 A0A182RVZ2 A0A182XN24 A0A182PTU9 A0A2J7RCP8 A0A1L8DUA4 V5RF49 A0A1W4X083 A0A336M8P4 A0A336MS14 T1PFF7 A0A1Q3FWH6 A0A182NBG3 T1E7V0 A0A182F260 A0A2M4BJV3 W5JEP1 E0VTG8 A0A0A1WR41 A0A1B0CV76 D3TMR1 A0A0L0CJW2 A0A034WLE7 A0A1J1IQ64 A0A088AAV5 F2D638 A0A0K8WEE3 A0A0K8U9A7 A0A1L8EDC8 A0A1I8P7C9 A0A1A9UEE5 A0A2H8U143 A0A2S2NU54 A0A0N0BHT9 A0A1L8EDQ5 A0A0K8TSS4 K7J2G8 J9K0Z9 A0A2A3EE83 W8BBM5 A0A1B0FGM9 A0A310SE98 A0A2M4A9Q8 A0A067RHG6 A0A2H8TFP5 A0A1B0DIB3 A0A1B0ADK9 F4WS67 A0A0M5JAW2 A0A158N9W9 A0A0L7QTR4 A0A1A9YS69 A0A151WRU9 A0A151ISP9 A0A182J824 A0A1A9X0B7 J9JK95 A0A0C9QE42 A0A0C9RYP3 J3JUX5 A0A1Q3FX16 N6U469 A0A3B0KWY2 B4M281 B4JIV9 A0A3L8E321 B3MZK0 A0A1W4UD59 E2BQ93 E2AE48 Q29GX0 B4IJQ6 B3NUH9 B4PZY2 B4N1Z4
A0A194Q8Z1 A0A182UXK1 A0A182TK59 A0A182LMA0 A0A182HI69 A0A1B6CS56 A0A1B6GB87 Q7Q1P2 A0A182QGJ2 D6WBW7 U5EZ38 A0A182YFT8 A0A182KFJ9 A0A1B6LBH7 A0A023ETP2 A0A1S4FHA0 V5GNR1 A0A1Y1L6K5 A0A182M9I4 A0A182VU05 A0A182RVZ2 A0A182XN24 A0A182PTU9 A0A2J7RCP8 A0A1L8DUA4 V5RF49 A0A1W4X083 A0A336M8P4 A0A336MS14 T1PFF7 A0A1Q3FWH6 A0A182NBG3 T1E7V0 A0A182F260 A0A2M4BJV3 W5JEP1 E0VTG8 A0A0A1WR41 A0A1B0CV76 D3TMR1 A0A0L0CJW2 A0A034WLE7 A0A1J1IQ64 A0A088AAV5 F2D638 A0A0K8WEE3 A0A0K8U9A7 A0A1L8EDC8 A0A1I8P7C9 A0A1A9UEE5 A0A2H8U143 A0A2S2NU54 A0A0N0BHT9 A0A1L8EDQ5 A0A0K8TSS4 K7J2G8 J9K0Z9 A0A2A3EE83 W8BBM5 A0A1B0FGM9 A0A310SE98 A0A2M4A9Q8 A0A067RHG6 A0A2H8TFP5 A0A1B0DIB3 A0A1B0ADK9 F4WS67 A0A0M5JAW2 A0A158N9W9 A0A0L7QTR4 A0A1A9YS69 A0A151WRU9 A0A151ISP9 A0A182J824 A0A1A9X0B7 J9JK95 A0A0C9QE42 A0A0C9RYP3 J3JUX5 A0A1Q3FX16 N6U469 A0A3B0KWY2 B4M281 B4JIV9 A0A3L8E321 B3MZK0 A0A1W4UD59 E2BQ93 E2AE48 Q29GX0 B4IJQ6 B3NUH9 B4PZY2 B4N1Z4
PDB
4B2T
E-value=1.98028e-175,
Score=1582
Ontologies
GO
PANTHER
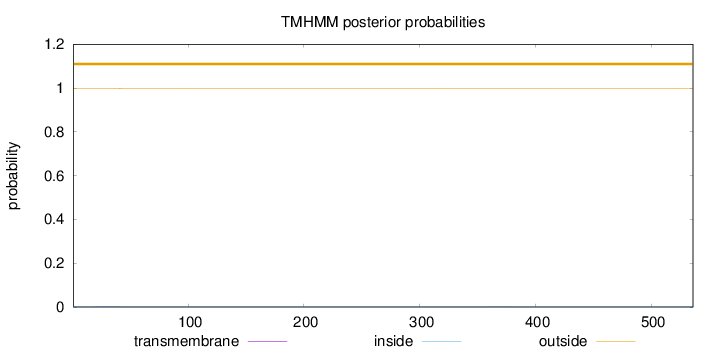
Topology
Subcellular location
Cytoplasm
Length:
536
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0512000000000001
Exp number, first 60 AAs:
0.03929
Total prob of N-in:
0.00253
outside
1 - 536
Population Genetic Test Statistics
Pi
279.908947
Theta
207.035787
Tajima's D
1.924217
CLR
0.149501
CSRT
0.868956552172391
Interpretation
Uncertain
