Gene
KWMTBOMO07854
Pre Gene Modal
BGIBMGA000941
Annotation
PREDICTED:_semaphorin-2A-like_isoform_X1_[Bombyx_mori]
Full name
Semaphorin-2A
Alternative Name
Sema II
Semaphorin-II
Semaphorin-II
Location in the cell
Extracellular Reliability : 1.859
Sequence
CDS
ATGAACGAAGAGAGAAATACGCTTTATGTTGGTGCTATGGATCGTGTATTCAAATTAAATATGTCTGATATTAGTCAATCACATTGTGAGAGAGATGCATTGTTATTAGAAGCTAGTAATGTAGCGAGATGTGTTAGTAAAGGAAAATCAGAGCCTTTTGAGTGTAGAAACCATATTCGTGTGCTGCAGCCGCTGGGCGACGGAGAGAGGCTGTACGTTTGCGGTACTAATGCACATAGCCCCAAAGATTGGGTCGTGTATTTAAATTTAACGCATCTACCTCGGCACGAGTATATACCGGGCGTGGGCCTTGGTGTGGCCAAATGTCCATACGACCCTGCGGACAATAGTACTGCAGTATGGGTCACCGAGGGCAATCCTGGGGAGCTTCCAGCTTTGTACGCTGGTACCAATGCAGAGTTCACGAAGGCTGACCCAGTTATATTTCGTAATGATCTCTTTGATTTTCGGACTGGTCAGAAGAAATTTAATTTTAAGAGAACATTAAAATATGATTCCAAGTGGTTGGATAAAACAAATTTCGTTGGTTCATTTGATGTGGGTGAATATGTTTACTTTTTCTTCCGCGAGACAGCGGTGGAGTTTATGAACTGTGGTAAAGCAGTATATTCAAGAGTAGCCAGAGTTTGTAAACAGGATAACGGAGGGAAACATATCCTCAGTCAAAATTGGGTCACTTATCTTAAAGCAAGACTTAATTGCAGCATACCTGGAGAATTTCCTTTTTATTTTGATGAAATACAAAGTGTACATCAAATACCGGGGGATACGAGCCGGTTTTACGCGGCGTTCACGACGGGCGCCAGTGATGGCCTCATAGGATCCGCGATATGCACTTACACTATGGACGATATACAGGAGGCTTTCGCGGGAAGATTCAAAGAACAGGCTTCGTCAAGTTCAGCATGGCTTCCAGTGCTGCGAGCTCGAGTGCCAGTGCCACGTCCCGGCACTTGCGTCAATAAAACCGAAGCTTTACCAGACAATGTCCTCAACTTTATAAGGTCTCACCCGTTGATGGATTCAGCTGTGCGACACGACGGTGATGCACCTGCTTTCTACAAGCGTGATCTGGTGCTCACCACTCTTGTAGTAGACAAACAGTTTGTTGACATGCTGGGAGATGACATCACGTACACAGTATTTTATGCCGGAACTAATGCAGGCGAAGTATATAAGATAGTGCAGTGGGCGGGAGGGGGCGCTCGTGTGGCTGATGTATGGCTGGCCGCCGCAGCAGAGCCTATTCGCGCCCTCGCACTCGCACGCACTCAACACGCGCTGTACCTAGCCACTGATCATAGATTAAGAGAACTACCACTAAGAGCTTGTGCTCATCGTTACGATAGCTGCGTGCGTTGTGTACTAGATCCGTACTGTGGTTGGGACAAAGAAGTAGGGGCGTGTCGTCCGTACACTCCCGGCCTACTTCACGACGCCACAAACTCAACTCCTTCGATATGCGAGGCTAGTGCCCCTCTGAAAACTGTGAAGGCGAGCTACGGCCAAAGCGTCCATTTAGCGGCCTTCGGGAAGACGCCGGATGCACTGAAAGACCAATACGTCGTATGGTTTCATCATTCCAGAGAGAAGGGACGGTACAAAATTAATTTCAACTGGGATCGAGTGCTACAAACATCGGAGCGAGGTCTCGTTCTACTCTCTGTGTCCGACTCGGACCGAGGCCGATACGAATGCTACTTTGGACCTTCTCTTGTGGCTTCATACAATCTAGATATTGATATACACAGGTGTACACAACCGAGTAAAACTCAAGAATATAAACAAGTGTATTCCGATTGGTGTCATGAGTTCGAGAAATACAAGAACGCTATGAAAGTATGGGAGTCACGGCAAATGCAATGCGCGAAGGGCATGAATGCATCGAGTCAACAGAACAGTATGAACAACGAGCTCGTTGCGTCGCTGCGGTGA
Protein
MNEERNTLYVGAMDRVFKLNMSDISQSHCERDALLLEASNVARCVSKGKSEPFECRNHIRVLQPLGDGERLYVCGTNAHSPKDWVVYLNLTHLPRHEYIPGVGLGVAKCPYDPADNSTAVWVTEGNPGELPALYAGTNAEFTKADPVIFRNDLFDFRTGQKKFNFKRTLKYDSKWLDKTNFVGSFDVGEYVYFFFRETAVEFMNCGKAVYSRVARVCKQDNGGKHILSQNWVTYLKARLNCSIPGEFPFYFDEIQSVHQIPGDTSRFYAAFTTGASDGLIGSAICTYTMDDIQEAFAGRFKEQASSSSAWLPVLRARVPVPRPGTCVNKTEALPDNVLNFIRSHPLMDSAVRHDGDAPAFYKRDLVLTTLVVDKQFVDMLGDDITYTVFYAGTNAGEVYKIVQWAGGGARVADVWLAAAAEPIRALALARTQHALYLATDHRLRELPLRACAHRYDSCVRCVLDPYCGWDKEVGACRPYTPGLLHDATNSTPSICEASAPLKTVKASYGQSVHLAAFGKTPDALKDQYVVWFHHSREKGRYKINFNWDRVLQTSERGLVLLSVSDSDRGRYECYFGPSLVASYNLDIDIHRCTQPSKTQEYKQVYSDWCHEFEKYKNAMKVWESRQMQCAKGMNASSQQNSMNNELVASLR
Summary
Description
Plays a role in growth cones guidance. Required for both proper adult behavior and survival. Can function in vivo as a selective target-derived signal that inhibits the formation of specific synaptic terminal arbors.
Subunit
Interacts with PlexB.
Similarity
Belongs to the semaphorin family.
Keywords
Developmental protein
Differentiation
Disulfide bond
Glycoprotein
Immunoglobulin domain
Neurogenesis
Secreted
Signal
Alternative splicing
Complete proteome
Reference proteome
Feature
chain Semaphorin-2A
splice variant In isoform C.
splice variant In isoform C.
Uniprot
A0A194QAG8
A0A194RH66
A0A3S2NF77
A0A2A4JA10
A0A3S2LY87
A0A194RRL5
+ More
D6BK68 K7J7T3 A0A195EDQ7 A0A158NY55 A0A088A483 A0A139WNC0 A0A139WNB6 A0A026WKJ4 A0MT98 A0A154NWA1 A0A1B6FNE2 Q7Q552 A0A1Y1NCB7 A0A1Y1NE22 A0A1Y1N840 A0A0C9RMB0 A0A1Y1N8R5 A0A1W7R6R0 A0A1B6C5M3 A0A0P6K0S6 A0A0A9Y8U9 A0A023EW03 A0A1Q3FWP5 Q177X8 A0A023F5G6 E0VMC9 A0A195B0N5 E2A5X9 A0A0P6GQ47 B4J7G7 A0A194PN26 A0A151IJF0 Q9XZC8 A0A3B0IZ98 B4MPH4 B4QI01 A0A1W4UYA5 A0A3B0JJQ7 A0A0R3NLI8 A0A0Q9XIE5 B4HSZ1 Q24323-2 B4LME9 A0A1W4UL16 A0A0J9U3N0 B4KSI2 C0PTV9 A0A0Q9WG98 B3NPH7 B5DZT9 B4P659 A0A0P9C0E2 Q24323 A0A151WPV4 A0A0M4ECX6 A0A084WMF0 A0A0Q5VN12 A0A0R1DSV2 A0A0K8W563 B3MFQ2 A0A0A1WSG7 A0A182V3Z2 A0A1J1INU3 A0A0P6I923 A0A0P5YB13 W8BL50 A0A0P5BDT7 A0A0P5ESR9 A0A0P6IH40 A0A0P5EVZ2 W8BB33 Q7PP64 A0A0P5A215 A0A0P5G8G5 A0A336MPW5 B0XD36 A0A0P5EKF9 A0A2H8TGV7 A0A1I8P3Q1 A0A0P5UPR1 J9K5D2 A0A1I8NGM9 A0A0P5TW35 A0A0N8CJF1 A0A2S2PAN5 Q24323-4 A0A0P5P7H7 A0A0P5P7F9 A0A0K8TZQ3 A0A0L0BLD8 A0A2P2HZ25 A0A067QIQ5 A0A182R981
D6BK68 K7J7T3 A0A195EDQ7 A0A158NY55 A0A088A483 A0A139WNC0 A0A139WNB6 A0A026WKJ4 A0MT98 A0A154NWA1 A0A1B6FNE2 Q7Q552 A0A1Y1NCB7 A0A1Y1NE22 A0A1Y1N840 A0A0C9RMB0 A0A1Y1N8R5 A0A1W7R6R0 A0A1B6C5M3 A0A0P6K0S6 A0A0A9Y8U9 A0A023EW03 A0A1Q3FWP5 Q177X8 A0A023F5G6 E0VMC9 A0A195B0N5 E2A5X9 A0A0P6GQ47 B4J7G7 A0A194PN26 A0A151IJF0 Q9XZC8 A0A3B0IZ98 B4MPH4 B4QI01 A0A1W4UYA5 A0A3B0JJQ7 A0A0R3NLI8 A0A0Q9XIE5 B4HSZ1 Q24323-2 B4LME9 A0A1W4UL16 A0A0J9U3N0 B4KSI2 C0PTV9 A0A0Q9WG98 B3NPH7 B5DZT9 B4P659 A0A0P9C0E2 Q24323 A0A151WPV4 A0A0M4ECX6 A0A084WMF0 A0A0Q5VN12 A0A0R1DSV2 A0A0K8W563 B3MFQ2 A0A0A1WSG7 A0A182V3Z2 A0A1J1INU3 A0A0P6I923 A0A0P5YB13 W8BL50 A0A0P5BDT7 A0A0P5ESR9 A0A0P6IH40 A0A0P5EVZ2 W8BB33 Q7PP64 A0A0P5A215 A0A0P5G8G5 A0A336MPW5 B0XD36 A0A0P5EKF9 A0A2H8TGV7 A0A1I8P3Q1 A0A0P5UPR1 J9K5D2 A0A1I8NGM9 A0A0P5TW35 A0A0N8CJF1 A0A2S2PAN5 Q24323-4 A0A0P5P7H7 A0A0P5P7F9 A0A0K8TZQ3 A0A0L0BLD8 A0A2P2HZ25 A0A067QIQ5 A0A182R981
Pubmed
26354079
20075255
21347285
18362917
19820115
24508170
+ More
12364791 28004739 26999592 25401762 24945155 17510324 25474469 20566863 20798317 17994087 10101134 22936249 15632085 8269517 10731132 12537572 12537569 16672342 19349973 18057021 17550304 24438588 25830018 24495485 25315136 26108605 24845553
12364791 28004739 26999592 25401762 24945155 17510324 25474469 20566863 20798317 17994087 10101134 22936249 15632085 8269517 10731132 12537572 12537569 16672342 19349973 18057021 17550304 24438588 25830018 24495485 25315136 26108605 24845553
EMBL
KQ459299
KPJ01990.1
KQ460205
KPJ16922.1
RSAL01000144
RVE45961.1
+ More
NWSH01002468 PCG68233.1 RSAL01000119 RVE46737.1 KQ459765 KPJ20039.1 EU836737 ACJ23177.1 AAZX01003580 AAZX01006728 AAZX01007460 AAZX01008558 AAZX01009344 AAZX01009609 AAZX01020005 KQ979074 KYN22979.1 ADTU01003207 ADTU01003208 ADTU01003209 ADTU01003210 ADTU01003211 ADTU01003212 KQ971312 KYB29357.1 KYB29356.1 KK107168 EZA56161.1 EF036538 ABK35089.3 KQ434772 KZC03927.1 GECZ01020839 GECZ01018246 JAS48930.1 JAS51523.1 AAAB01008960 EAA11400.4 GEZM01010727 JAV93876.1 GEZM01010726 JAV93877.1 GEZM01010724 JAV93879.1 GBYB01009445 JAG79212.1 GEZM01010725 JAV93878.1 GEHC01000845 JAV46800.1 GEDC01028490 JAS08808.1 GDUN01000846 JAN95073.1 GBHO01015025 GBRD01015533 JAG28579.1 JAG50293.1 GAPW01000322 JAC13276.1 GFDL01003077 JAV31968.1 CH477369 EAT42479.1 GBBI01002318 JAC16394.1 DS235306 EEB14535.1 KQ976692 KYM77764.1 GL437072 EFN71160.1 GDIQ01040643 JAN54094.1 CH916367 EDW01091.1 KQ459604 KPI92525.1 KQ977355 KYN03331.1 AF134904 OUUW01000001 SPP73455.1 CH963849 EDW74013.1 CM000362 CM002911 EDX07372.1 KMY94279.1 SPP73456.1 CM000071 KRT01800.1 CH933808 KRG04686.1 CH480816 EDW48155.1 L26083 AE013599 AY095026 AY128437 BT011447 CH940648 EDW62044.2 KMY94280.1 KMY94281.1 KMY94282.1 EDW09487.1 BT071804 ACN43740.1 KRF80399.1 KRF80400.1 CH954179 EDV55744.1 EDY68862.1 CM000158 EDW91909.1 CH902619 KPU77008.1 KQ982851 KYQ49896.1 CP012524 ALC41677.1 ATLV01024426 ATLV01024427 KE525352 KFB51394.1 KQS62451.1 KQS62452.1 KRK00203.1 KRK00204.1 KRK00205.1 GDHF01034064 GDHF01027113 GDHF01019851 GDHF01006112 GDHF01002618 JAI18250.1 JAI25201.1 JAI32463.1 JAI46202.1 JAI49696.1 EDV37742.1 KPU77009.1 KPU77010.1 GBXI01012515 JAD01777.1 CVRI01000054 CRL00153.1 GDIQ01024602 JAN70135.1 GDIP01062188 JAM41527.1 GAMC01016191 JAB90364.1 GDIP01186221 GDIQ01116330 GDIQ01059403 JAJ37181.1 JAL35396.1 GDIP01174818 GDIQ01268439 GDIQ01106037 GDIQ01059404 GDIQ01049163 LRGB01002993 JAJ48584.1 JAJ83285.1 KZS05205.1 GDIQ01004618 JAN90119.1 GDIQ01266422 JAJ85302.1 GAMC01016189 GAMC01016188 JAB90366.1 EAA11003.5 GDIP01217514 JAJ05888.1 GDIQ01266421 JAJ85303.1 UFQS01002077 UFQT01002077 SSX13155.1 SSX32594.1 DS232737 EDS45271.1 GDIQ01268438 JAJ83286.1 GFXV01001551 MBW13356.1 GDIP01111340 JAL92374.1 ABLF02036314 ABLF02036316 ABLF02036319 ABLF02036320 ABLF02036322 ABLF02036323 ABLF02047241 ABLF02047629 ABLF02048663 ABLF02066977 GDIP01125889 JAL77825.1 GDIP01125839 JAL77875.1 GGMR01013377 MBY25996.1 GDIQ01132581 JAL19145.1 GDIQ01132582 JAL19144.1 GDHF01032367 GDHF01022494 JAI19947.1 JAI29820.1 JRES01001695 KNC20905.1 IACF01001160 LAB66876.1 KK853319 KDR08559.1
NWSH01002468 PCG68233.1 RSAL01000119 RVE46737.1 KQ459765 KPJ20039.1 EU836737 ACJ23177.1 AAZX01003580 AAZX01006728 AAZX01007460 AAZX01008558 AAZX01009344 AAZX01009609 AAZX01020005 KQ979074 KYN22979.1 ADTU01003207 ADTU01003208 ADTU01003209 ADTU01003210 ADTU01003211 ADTU01003212 KQ971312 KYB29357.1 KYB29356.1 KK107168 EZA56161.1 EF036538 ABK35089.3 KQ434772 KZC03927.1 GECZ01020839 GECZ01018246 JAS48930.1 JAS51523.1 AAAB01008960 EAA11400.4 GEZM01010727 JAV93876.1 GEZM01010726 JAV93877.1 GEZM01010724 JAV93879.1 GBYB01009445 JAG79212.1 GEZM01010725 JAV93878.1 GEHC01000845 JAV46800.1 GEDC01028490 JAS08808.1 GDUN01000846 JAN95073.1 GBHO01015025 GBRD01015533 JAG28579.1 JAG50293.1 GAPW01000322 JAC13276.1 GFDL01003077 JAV31968.1 CH477369 EAT42479.1 GBBI01002318 JAC16394.1 DS235306 EEB14535.1 KQ976692 KYM77764.1 GL437072 EFN71160.1 GDIQ01040643 JAN54094.1 CH916367 EDW01091.1 KQ459604 KPI92525.1 KQ977355 KYN03331.1 AF134904 OUUW01000001 SPP73455.1 CH963849 EDW74013.1 CM000362 CM002911 EDX07372.1 KMY94279.1 SPP73456.1 CM000071 KRT01800.1 CH933808 KRG04686.1 CH480816 EDW48155.1 L26083 AE013599 AY095026 AY128437 BT011447 CH940648 EDW62044.2 KMY94280.1 KMY94281.1 KMY94282.1 EDW09487.1 BT071804 ACN43740.1 KRF80399.1 KRF80400.1 CH954179 EDV55744.1 EDY68862.1 CM000158 EDW91909.1 CH902619 KPU77008.1 KQ982851 KYQ49896.1 CP012524 ALC41677.1 ATLV01024426 ATLV01024427 KE525352 KFB51394.1 KQS62451.1 KQS62452.1 KRK00203.1 KRK00204.1 KRK00205.1 GDHF01034064 GDHF01027113 GDHF01019851 GDHF01006112 GDHF01002618 JAI18250.1 JAI25201.1 JAI32463.1 JAI46202.1 JAI49696.1 EDV37742.1 KPU77009.1 KPU77010.1 GBXI01012515 JAD01777.1 CVRI01000054 CRL00153.1 GDIQ01024602 JAN70135.1 GDIP01062188 JAM41527.1 GAMC01016191 JAB90364.1 GDIP01186221 GDIQ01116330 GDIQ01059403 JAJ37181.1 JAL35396.1 GDIP01174818 GDIQ01268439 GDIQ01106037 GDIQ01059404 GDIQ01049163 LRGB01002993 JAJ48584.1 JAJ83285.1 KZS05205.1 GDIQ01004618 JAN90119.1 GDIQ01266422 JAJ85302.1 GAMC01016189 GAMC01016188 JAB90366.1 EAA11003.5 GDIP01217514 JAJ05888.1 GDIQ01266421 JAJ85303.1 UFQS01002077 UFQT01002077 SSX13155.1 SSX32594.1 DS232737 EDS45271.1 GDIQ01268438 JAJ83286.1 GFXV01001551 MBW13356.1 GDIP01111340 JAL92374.1 ABLF02036314 ABLF02036316 ABLF02036319 ABLF02036320 ABLF02036322 ABLF02036323 ABLF02047241 ABLF02047629 ABLF02048663 ABLF02066977 GDIP01125889 JAL77825.1 GDIP01125839 JAL77875.1 GGMR01013377 MBY25996.1 GDIQ01132581 JAL19145.1 GDIQ01132582 JAL19144.1 GDHF01032367 GDHF01022494 JAI19947.1 JAI29820.1 JRES01001695 KNC20905.1 IACF01001160 LAB66876.1 KK853319 KDR08559.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000218220
UP000002358
UP000078492
+ More
UP000005205 UP000005203 UP000007266 UP000053097 UP000076502 UP000007062 UP000008820 UP000009046 UP000078540 UP000000311 UP000001070 UP000078542 UP000268350 UP000007798 UP000000304 UP000192221 UP000001819 UP000009192 UP000001292 UP000000803 UP000008792 UP000008711 UP000002282 UP000007801 UP000075809 UP000092553 UP000030765 UP000075903 UP000183832 UP000076858 UP000002320 UP000095300 UP000007819 UP000095301 UP000037069 UP000027135 UP000075900
UP000005205 UP000005203 UP000007266 UP000053097 UP000076502 UP000007062 UP000008820 UP000009046 UP000078540 UP000000311 UP000001070 UP000078542 UP000268350 UP000007798 UP000000304 UP000192221 UP000001819 UP000009192 UP000001292 UP000000803 UP000008792 UP000008711 UP000002282 UP000007801 UP000075809 UP000092553 UP000030765 UP000075903 UP000183832 UP000076858 UP000002320 UP000095300 UP000007819 UP000095301 UP000037069 UP000027135 UP000075900
Interpro
Gene 3D
ProteinModelPortal
A0A194QAG8
A0A194RH66
A0A3S2NF77
A0A2A4JA10
A0A3S2LY87
A0A194RRL5
+ More
D6BK68 K7J7T3 A0A195EDQ7 A0A158NY55 A0A088A483 A0A139WNC0 A0A139WNB6 A0A026WKJ4 A0MT98 A0A154NWA1 A0A1B6FNE2 Q7Q552 A0A1Y1NCB7 A0A1Y1NE22 A0A1Y1N840 A0A0C9RMB0 A0A1Y1N8R5 A0A1W7R6R0 A0A1B6C5M3 A0A0P6K0S6 A0A0A9Y8U9 A0A023EW03 A0A1Q3FWP5 Q177X8 A0A023F5G6 E0VMC9 A0A195B0N5 E2A5X9 A0A0P6GQ47 B4J7G7 A0A194PN26 A0A151IJF0 Q9XZC8 A0A3B0IZ98 B4MPH4 B4QI01 A0A1W4UYA5 A0A3B0JJQ7 A0A0R3NLI8 A0A0Q9XIE5 B4HSZ1 Q24323-2 B4LME9 A0A1W4UL16 A0A0J9U3N0 B4KSI2 C0PTV9 A0A0Q9WG98 B3NPH7 B5DZT9 B4P659 A0A0P9C0E2 Q24323 A0A151WPV4 A0A0M4ECX6 A0A084WMF0 A0A0Q5VN12 A0A0R1DSV2 A0A0K8W563 B3MFQ2 A0A0A1WSG7 A0A182V3Z2 A0A1J1INU3 A0A0P6I923 A0A0P5YB13 W8BL50 A0A0P5BDT7 A0A0P5ESR9 A0A0P6IH40 A0A0P5EVZ2 W8BB33 Q7PP64 A0A0P5A215 A0A0P5G8G5 A0A336MPW5 B0XD36 A0A0P5EKF9 A0A2H8TGV7 A0A1I8P3Q1 A0A0P5UPR1 J9K5D2 A0A1I8NGM9 A0A0P5TW35 A0A0N8CJF1 A0A2S2PAN5 Q24323-4 A0A0P5P7H7 A0A0P5P7F9 A0A0K8TZQ3 A0A0L0BLD8 A0A2P2HZ25 A0A067QIQ5 A0A182R981
D6BK68 K7J7T3 A0A195EDQ7 A0A158NY55 A0A088A483 A0A139WNC0 A0A139WNB6 A0A026WKJ4 A0MT98 A0A154NWA1 A0A1B6FNE2 Q7Q552 A0A1Y1NCB7 A0A1Y1NE22 A0A1Y1N840 A0A0C9RMB0 A0A1Y1N8R5 A0A1W7R6R0 A0A1B6C5M3 A0A0P6K0S6 A0A0A9Y8U9 A0A023EW03 A0A1Q3FWP5 Q177X8 A0A023F5G6 E0VMC9 A0A195B0N5 E2A5X9 A0A0P6GQ47 B4J7G7 A0A194PN26 A0A151IJF0 Q9XZC8 A0A3B0IZ98 B4MPH4 B4QI01 A0A1W4UYA5 A0A3B0JJQ7 A0A0R3NLI8 A0A0Q9XIE5 B4HSZ1 Q24323-2 B4LME9 A0A1W4UL16 A0A0J9U3N0 B4KSI2 C0PTV9 A0A0Q9WG98 B3NPH7 B5DZT9 B4P659 A0A0P9C0E2 Q24323 A0A151WPV4 A0A0M4ECX6 A0A084WMF0 A0A0Q5VN12 A0A0R1DSV2 A0A0K8W563 B3MFQ2 A0A0A1WSG7 A0A182V3Z2 A0A1J1INU3 A0A0P6I923 A0A0P5YB13 W8BL50 A0A0P5BDT7 A0A0P5ESR9 A0A0P6IH40 A0A0P5EVZ2 W8BB33 Q7PP64 A0A0P5A215 A0A0P5G8G5 A0A336MPW5 B0XD36 A0A0P5EKF9 A0A2H8TGV7 A0A1I8P3Q1 A0A0P5UPR1 J9K5D2 A0A1I8NGM9 A0A0P5TW35 A0A0N8CJF1 A0A2S2PAN5 Q24323-4 A0A0P5P7H7 A0A0P5P7F9 A0A0K8TZQ3 A0A0L0BLD8 A0A2P2HZ25 A0A067QIQ5 A0A182R981
PDB
6FKN
E-value=1.88959e-67,
Score=652
Ontologies
GO
GO:0050919
GO:0071526
GO:0030335
GO:0045499
GO:0005615
GO:0007411
GO:0001755
GO:0048843
GO:0030215
GO:0016020
GO:0007399
GO:0030154
GO:0005576
GO:0007632
GO:0070983
GO:0016201
GO:0007629
GO:0042756
GO:0071678
GO:0097374
GO:0016021
GO:0030534
GO:0012505
GO:0005829
GO:0005515
GO:0016758
GO:0008270
GO:0005506
GO:0030145
GO:0050906
GO:0016818
GO:0003707
GO:0051537
PANTHER
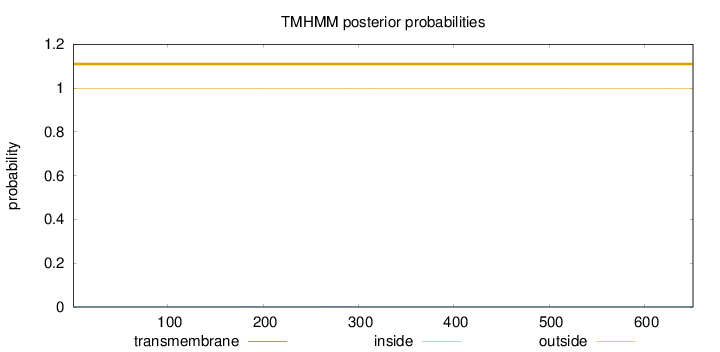
Topology
Subcellular location
Secreted
Length:
651
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02494
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00192
outside
1 - 651
Population Genetic Test Statistics
Pi
251.715206
Theta
181.240965
Tajima's D
1.186722
CLR
0.181947
CSRT
0.71766411679416
Interpretation
Uncertain
