Gene
KWMTBOMO07851
Pre Gene Modal
BGIBMGA000938
Annotation
PREDICTED:_probable_dolichyl_pyrophosphate_Glc1Man9GlcNAc2_alpha-1?3-glucosyltransferase_[Amyelois_transitella]
Full name
Alpha-1,3-glucosyltransferase
+ More
Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase
Probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase
Alternative Name
Asparagine-linked glycosylation protein 8 homolog
Dol-P-Glc:Glc(1)Man(9)GlcNAc(2)-PP-dolichyl alpha-1,3-glucosyltransferase
Dolichyl-P-Glc:Glc1Man9GlcNAc2-PP-dolichyl glucosyltransferase
Dol-P-Glc:Glc(1)Man(9)GlcNAc(2)-PP-dolichyl alpha-1,3-glucosyltransferase
Dolichyl-P-Glc:Glc1Man9GlcNAc2-PP-dolichyl glucosyltransferase
Location in the cell
PlasmaMembrane Reliability : 4.98
Sequence
CDS
ATGATTACCCAAGTAGTACTAATAGTTACAGCCGTTAAGTTTATGTTCATACCATTGTATCATTCAACAGACTTTGAAGTGCATCGAAATTGGTTGGCTATCACGAATAATGTTTCCGTCGAAGAGTGGTATTACGAATCGACATCAGAGTGGACACTGGACTACCCTCCACTCTTTGCCTGGCTAGAATATGGCTTGTCTATTGTAGCAAAATATTTTGATCCCAAAATGATAAAAATAGAAAATCTAAATTATAAATCTGATATGGCCGTGCTATTCCAAAGATTATCTGTGATTTTTTTAGACTTTGTTTACATTTTTAGTGTTAAAAGTTGCTCAAATCTAGTCAGCAATGGAAAGCTATTAGTTTTTATTTTATTAGTGTCCAACCCTGGCTTGCTGATGGTTGATCATATACACTTTCAATATAATGGATTTTTATATGGTTTTCTTCTGTTTTCTATATCGAACATGATAAGGTGTAATCATCTACAAGCAGCATTCTGGTTTGCTATGTTACTTAACTTTAAACATATATTTTTATATATTGCCCCAGTGTATGTGGTATATTTGTTAAGAGCATACTGTTTCACTCTTTCATCAATAGACGGAGTTCACACAGCTTGGTATTCCTTTTCAATAACAAATTTTATTAAACTGGCATTTGTTGTTGTATCAGTATTTACTTTATCGTTTGGACCATTTTTAGACCATTTGAATCAGATTTTAGTCAGATTATTTCCATTTAAAAGAGGACTTTGCCACGCATATTGGGCTCCCAATTTCTGGGCTCTGTACAATTTAGTAACAAAATATGGAATTAACATACCGAAATCGGAAGCATCGATGACAGGAGGGCTAGTGCAAGAATACGACCATTCAGTTTTACCATCAATTACGCCGAGTTTCACTTTTATAATGACGTGTTTGTCTATGATGCCCGCATTGTACAAGATGTGGTGTCTTTGTGCGGACAGAAGATACAGACCTATTTGTTTTATAAGATGTATTGTCGTCTGTGCGACTTGTTCGTTTATGCTGGGTTGGCATGTTCACGAGAAAGCAATTTTGATGATTATAATACCGTTGAGTTTTCTGTCGGTTCTTGGCGACGTGGACGGAAGATTATTCATTTTACTCTCAACCGTTCAATCGTTCGACTCATCGAGTTGGAAGTTTGAAAATAAAAGTGAAATGGAGCGGAAGGAGTTGTTCATTCAGATTGGTGATTTGGCATTTTATTACCTTTAG
Protein
MITQVVLIVTAVKFMFIPLYHSTDFEVHRNWLAITNNVSVEEWYYESTSEWTLDYPPLFAWLEYGLSIVAKYFDPKMIKIENLNYKSDMAVLFQRLSVIFLDFVYIFSVKSCSNLVSNGKLLVFILLVSNPGLLMVDHIHFQYNGFLYGFLLFSISNMIRCNHLQAAFWFAMLLNFKHIFLYIAPVYVVYLLRAYCFTLSSIDGVHTAWYSFSITNFIKLAFVVVSVFTLSFGPFLDHLNQILVRLFPFKRGLCHAYWAPNFWALYNLVTKYGINIPKSEASMTGGLVQEYDHSVLPSITPSFTFIMTCLSMMPALYKMWCLCADRRYRPICFIRCIVVCATCSFMLGWHVHEKAILMIIIPLSFLSVLGDVDGRLFILLSTVQSFDSSSWKFENKSEMERKELFIQIGDLAFYYL
Summary
Description
Adds the second glucose residue to the lipid-linked oligosaccharide precursor for N-linked glycosylation. Transfers glucose from dolichyl phosphate glucose (Dol-P-Glc) onto the lipid-linked oligosaccharide Glc(1)Man(9)GlcNAc(2)-PP-Dol before it is transferred to the nascent peptide (By similarity). Required for PKD1/Polycystin-1 maturation and localization to the plasma membrane of the primary cilia (By similarity).
Catalytic Activity
alpha-D-Glc-(1->3)-alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-alpha-D-GlcNAc-diphosphodolichol + dolichyl beta-D-glucosyl phosphate = alpha-D-Glc-(1->3)-alpha-D-Glc-(1->3)-alpha-D-Man-(1->2)-alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->3)-[alpha-D-Man-(1->2)-alpha-D-Man-(1->6)]-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-alpha-D-GlcNAc-diphosphodolichol + dolichyl phosphate + H(+)
Similarity
Belongs to the ALG6/ALG8 glucosyltransferase family.
Keywords
Alternative splicing
Complete proteome
Congenital disorder of glycosylation
Disease mutation
Endoplasmic reticulum
Glycosyltransferase
Membrane
Polymorphism
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain Alpha-1,3-glucosyltransferase
splice variant In isoform 2.
sequence variant In CDG1H; dbSNP:rs121908293.
splice variant In isoform 2.
sequence variant In CDG1H; dbSNP:rs121908293.
Uniprot
A0A1E1WIM4
M4M269
A0A2H1VGR7
A0A2A4JTX3
A0A212ELX4
A0A1B6FV48
+ More
A0A2J7PPT6 A0A1B6HEF3 A0A1W4WBF7 A0A1A7YLK7 A0A1B6BXI8 A0A1A8QQS1 W5KCM7 A0A1A8DXA2 A0A1A8FHY1 A0A1A8BZ40 A0A1S3KH68 A0A3Q1I1Z5 A0A1A8GX15 A0A3P8XWD8 A0A1A8KEN3 A0A1A8QZ63 Q6ZM97 Q566T2 A0A1W4ZUI4 A0A210QT88 A0A3B4CED4 A0A3B4WNL1 A0A1A8ADB6 A0A1S3HMK8 A0A3B3T263 A0A1B6D306 A0A1L8HB15 A0A3P8XW97 A0A3Q3NAA3 A0A1B6M1M6 A0A3B4VPT0 A0A3B5A2Y4 F7A881 A0A2U9B6Y2 Q5FVB1 A4IHH8 A0A2J8V0Q0 W5UBQ7 A0A3Q1FP80 L8HVM2 Q9BVK2-2 A0A2I2ZXK9 A0A2K6C3K5 A0A2K5TXY9 A0A2D0SYZ3 A0A2J8MBN0 A0A2K5J2Y7 A0A2K5P155 A0A2I3LFC5 A0A067RF74 A0A2K6QEG2 M3ZBQ1 A0A3Q0TH41 F1MV61 A0A3P8TYA6 A0A1S3RG10 Q0P5D9 A0A3B3DQC3 A0A3P8ZTD2 A0A0F8APD9 A0A2J8V0P2 A0A2K6C3F4 K7ARD9 Q6NRA2 G2HE43 A0A024R5K5 Q9BVK2 H2Q4H1 F6WKY0 G3R7I9 A0A2K6C3I2 G7PNB7 H9EVM6 A0A2R9ABB6 A0A2K5TXV6 A0A3Q7T9G9 K9KBB2 F6VZR9 A0A2I4CUA4 A0A2K5J2R6 A0A3Q3DS33 A0A096MVL3 A0A2K5P150 A0A2Y9KDL1 U6DG13 W5P9G0 A0A2I3HB56 A0A3Q3JGS2 A0A2K6QEE8 M3WZ59 I3L8Z7 A0A0D9QUY0 A0A3P9IC15 A0A2I3T0N0
A0A2J7PPT6 A0A1B6HEF3 A0A1W4WBF7 A0A1A7YLK7 A0A1B6BXI8 A0A1A8QQS1 W5KCM7 A0A1A8DXA2 A0A1A8FHY1 A0A1A8BZ40 A0A1S3KH68 A0A3Q1I1Z5 A0A1A8GX15 A0A3P8XWD8 A0A1A8KEN3 A0A1A8QZ63 Q6ZM97 Q566T2 A0A1W4ZUI4 A0A210QT88 A0A3B4CED4 A0A3B4WNL1 A0A1A8ADB6 A0A1S3HMK8 A0A3B3T263 A0A1B6D306 A0A1L8HB15 A0A3P8XW97 A0A3Q3NAA3 A0A1B6M1M6 A0A3B4VPT0 A0A3B5A2Y4 F7A881 A0A2U9B6Y2 Q5FVB1 A4IHH8 A0A2J8V0Q0 W5UBQ7 A0A3Q1FP80 L8HVM2 Q9BVK2-2 A0A2I2ZXK9 A0A2K6C3K5 A0A2K5TXY9 A0A2D0SYZ3 A0A2J8MBN0 A0A2K5J2Y7 A0A2K5P155 A0A2I3LFC5 A0A067RF74 A0A2K6QEG2 M3ZBQ1 A0A3Q0TH41 F1MV61 A0A3P8TYA6 A0A1S3RG10 Q0P5D9 A0A3B3DQC3 A0A3P8ZTD2 A0A0F8APD9 A0A2J8V0P2 A0A2K6C3F4 K7ARD9 Q6NRA2 G2HE43 A0A024R5K5 Q9BVK2 H2Q4H1 F6WKY0 G3R7I9 A0A2K6C3I2 G7PNB7 H9EVM6 A0A2R9ABB6 A0A2K5TXV6 A0A3Q7T9G9 K9KBB2 F6VZR9 A0A2I4CUA4 A0A2K5J2R6 A0A3Q3DS33 A0A096MVL3 A0A2K5P150 A0A2Y9KDL1 U6DG13 W5P9G0 A0A2I3HB56 A0A3Q3JGS2 A0A2K6QEE8 M3WZ59 I3L8Z7 A0A0D9QUY0 A0A3P9IC15 A0A2I3T0N0
EC Number
2.4.1.-
2.4.1.265
2.4.1.265
Pubmed
22118469
25329095
25069045
23594743
28812685
29240929
+ More
27762356 17431167 23127152 22751099 11124703 16554811 15489334 12480927 15235028 28375157 22398555 16136131 24845553 25362486 19393038 29451363 25835551 21484476 11181995 26871637 17495919 22002653 25319552 22722832 19892987 20809919 17975172 30723633 17554307
27762356 17431167 23127152 22751099 11124703 16554811 15489334 12480927 15235028 28375157 22398555 16136131 24845553 25362486 19393038 29451363 25835551 21484476 11181995 26871637 17495919 22002653 25319552 22722832 19892987 20809919 17975172 30723633 17554307
EMBL
GDQN01004195
JAT86859.1
JQ747501
AGG36458.1
ODYU01002495
SOQ40043.1
+ More
NWSH01000642 PCG75074.1 AGBW02013971 OWR42492.1 GECZ01015701 JAS54068.1 NEVH01022644 PNF18357.1 GECU01034650 GECU01026325 JAS73056.1 JAS81381.1 HADW01002224 HADX01008573 SBP30805.1 GEDC01031569 JAS05729.1 HAEI01006045 SBR95891.1 HAEA01009406 SBQ37886.1 HAEB01011906 SBQ58433.1 HADZ01007925 SBP71866.1 HAEC01007476 SBQ75614.1 HAED01010746 HAEE01010742 SBR30792.1 HAEG01014900 SBR98802.1 AL627325 CAE47759.1 CU928848 BC093349 BC164687 AAH93349.1 AAI64687.1 NEDP02002015 OWF51951.1 HADY01013735 HAEJ01017962 SBP52220.1 GEDC01017272 JAS20026.1 CM004469 OCT93293.1 GEBQ01010154 GEBQ01007057 JAT29823.1 JAT32920.1 JSUE03012997 JSUE03012998 CP026246 AWO99595.1 BC090105 AAH90105.1 BC135531 AAI35532.1 ABGA01113927 ABGA01113928 ABGA01113929 ABGA01113930 ABGA01113931 ABGA01113932 NDHI03003437 PNJ51094.1 JT411033 AHH39285.1 JH883095 ELR47374.1 AJ224875 AP002520 CH471076 BC001133 CABD030080370 CABD030080371 CABD030080372 CABD030080373 AQIA01021535 AQIA01021536 AACZ04016310 NBAG03000261 PNI56929.1 AHZZ02007190 KK852672 KDR18773.1 ADFV01111229 ADFV01111230 ADFV01111231 ADFV01111232 ADFV01111233 ADFV01111234 ADFV01111235 ADFV01111236 BC120179 KQ041556 KKF25557.1 PNJ51085.1 GABC01006954 JAA04384.1 BC070861 AAH70861.1 AK305007 BAK62001.1 KU178764 ALQ34222.1 EAW75051.1 GABF01010490 GABD01004338 GABE01006028 JAA11655.1 JAA28762.1 JAA38711.1 PNI56926.1 CM001289 EHH56612.1 JU322679 JU472505 JV044589 AFE66435.1 AFH29309.1 AFI34660.1 AJFE02013302 AJFE02013303 AJFE02013304 AJFE02013305 AJFE02013306 AJFE02013307 JL620308 AEP99295.1 HAAF01009009 CCP80833.1 AMGL01060851 AANG04002890 AEMK02000070 DQIR01105371 DQIR01122511 DQIR01144885 DQIR01168884 DQIR01190177 DQIR01190989 DQIR01191825 DQIR01196356 DQIR01200500 DQIR01250151 DQIR01293354 HDA60847.1 AQIB01134049 AQIB01134050 AQIB01134051
NWSH01000642 PCG75074.1 AGBW02013971 OWR42492.1 GECZ01015701 JAS54068.1 NEVH01022644 PNF18357.1 GECU01034650 GECU01026325 JAS73056.1 JAS81381.1 HADW01002224 HADX01008573 SBP30805.1 GEDC01031569 JAS05729.1 HAEI01006045 SBR95891.1 HAEA01009406 SBQ37886.1 HAEB01011906 SBQ58433.1 HADZ01007925 SBP71866.1 HAEC01007476 SBQ75614.1 HAED01010746 HAEE01010742 SBR30792.1 HAEG01014900 SBR98802.1 AL627325 CAE47759.1 CU928848 BC093349 BC164687 AAH93349.1 AAI64687.1 NEDP02002015 OWF51951.1 HADY01013735 HAEJ01017962 SBP52220.1 GEDC01017272 JAS20026.1 CM004469 OCT93293.1 GEBQ01010154 GEBQ01007057 JAT29823.1 JAT32920.1 JSUE03012997 JSUE03012998 CP026246 AWO99595.1 BC090105 AAH90105.1 BC135531 AAI35532.1 ABGA01113927 ABGA01113928 ABGA01113929 ABGA01113930 ABGA01113931 ABGA01113932 NDHI03003437 PNJ51094.1 JT411033 AHH39285.1 JH883095 ELR47374.1 AJ224875 AP002520 CH471076 BC001133 CABD030080370 CABD030080371 CABD030080372 CABD030080373 AQIA01021535 AQIA01021536 AACZ04016310 NBAG03000261 PNI56929.1 AHZZ02007190 KK852672 KDR18773.1 ADFV01111229 ADFV01111230 ADFV01111231 ADFV01111232 ADFV01111233 ADFV01111234 ADFV01111235 ADFV01111236 BC120179 KQ041556 KKF25557.1 PNJ51085.1 GABC01006954 JAA04384.1 BC070861 AAH70861.1 AK305007 BAK62001.1 KU178764 ALQ34222.1 EAW75051.1 GABF01010490 GABD01004338 GABE01006028 JAA11655.1 JAA28762.1 JAA38711.1 PNI56926.1 CM001289 EHH56612.1 JU322679 JU472505 JV044589 AFE66435.1 AFH29309.1 AFI34660.1 AJFE02013302 AJFE02013303 AJFE02013304 AJFE02013305 AJFE02013306 AJFE02013307 JL620308 AEP99295.1 HAAF01009009 CCP80833.1 AMGL01060851 AANG04002890 AEMK02000070 DQIR01105371 DQIR01122511 DQIR01144885 DQIR01168884 DQIR01190177 DQIR01190989 DQIR01191825 DQIR01196356 DQIR01200500 DQIR01250151 DQIR01293354 HDA60847.1 AQIB01134049 AQIB01134050 AQIB01134051
Proteomes
UP000218220
UP000007151
UP000235965
UP000192223
UP000018467
UP000085678
+ More
UP000265040 UP000265140 UP000000437 UP000192224 UP000242188 UP000261440 UP000261360 UP000261540 UP000186698 UP000261640 UP000261420 UP000261400 UP000006718 UP000246464 UP000001595 UP000257200 UP000005640 UP000001519 UP000233120 UP000233100 UP000221080 UP000002277 UP000233080 UP000233060 UP000028761 UP000027135 UP000233200 UP000001073 UP000261340 UP000009136 UP000265080 UP000087266 UP000261560 UP000002280 UP000009130 UP000240080 UP000286640 UP000002281 UP000192220 UP000264820 UP000248482 UP000002356 UP000261600 UP000011712 UP000008227 UP000029965 UP000265200
UP000265040 UP000265140 UP000000437 UP000192224 UP000242188 UP000261440 UP000261360 UP000261540 UP000186698 UP000261640 UP000261420 UP000261400 UP000006718 UP000246464 UP000001595 UP000257200 UP000005640 UP000001519 UP000233120 UP000233100 UP000221080 UP000002277 UP000233080 UP000233060 UP000028761 UP000027135 UP000233200 UP000001073 UP000261340 UP000009136 UP000265080 UP000087266 UP000261560 UP000002280 UP000009130 UP000240080 UP000286640 UP000002281 UP000192220 UP000264820 UP000248482 UP000002356 UP000261600 UP000011712 UP000008227 UP000029965 UP000265200
Pfam
PF03155 Alg6_Alg8
ProteinModelPortal
A0A1E1WIM4
M4M269
A0A2H1VGR7
A0A2A4JTX3
A0A212ELX4
A0A1B6FV48
+ More
A0A2J7PPT6 A0A1B6HEF3 A0A1W4WBF7 A0A1A7YLK7 A0A1B6BXI8 A0A1A8QQS1 W5KCM7 A0A1A8DXA2 A0A1A8FHY1 A0A1A8BZ40 A0A1S3KH68 A0A3Q1I1Z5 A0A1A8GX15 A0A3P8XWD8 A0A1A8KEN3 A0A1A8QZ63 Q6ZM97 Q566T2 A0A1W4ZUI4 A0A210QT88 A0A3B4CED4 A0A3B4WNL1 A0A1A8ADB6 A0A1S3HMK8 A0A3B3T263 A0A1B6D306 A0A1L8HB15 A0A3P8XW97 A0A3Q3NAA3 A0A1B6M1M6 A0A3B4VPT0 A0A3B5A2Y4 F7A881 A0A2U9B6Y2 Q5FVB1 A4IHH8 A0A2J8V0Q0 W5UBQ7 A0A3Q1FP80 L8HVM2 Q9BVK2-2 A0A2I2ZXK9 A0A2K6C3K5 A0A2K5TXY9 A0A2D0SYZ3 A0A2J8MBN0 A0A2K5J2Y7 A0A2K5P155 A0A2I3LFC5 A0A067RF74 A0A2K6QEG2 M3ZBQ1 A0A3Q0TH41 F1MV61 A0A3P8TYA6 A0A1S3RG10 Q0P5D9 A0A3B3DQC3 A0A3P8ZTD2 A0A0F8APD9 A0A2J8V0P2 A0A2K6C3F4 K7ARD9 Q6NRA2 G2HE43 A0A024R5K5 Q9BVK2 H2Q4H1 F6WKY0 G3R7I9 A0A2K6C3I2 G7PNB7 H9EVM6 A0A2R9ABB6 A0A2K5TXV6 A0A3Q7T9G9 K9KBB2 F6VZR9 A0A2I4CUA4 A0A2K5J2R6 A0A3Q3DS33 A0A096MVL3 A0A2K5P150 A0A2Y9KDL1 U6DG13 W5P9G0 A0A2I3HB56 A0A3Q3JGS2 A0A2K6QEE8 M3WZ59 I3L8Z7 A0A0D9QUY0 A0A3P9IC15 A0A2I3T0N0
A0A2J7PPT6 A0A1B6HEF3 A0A1W4WBF7 A0A1A7YLK7 A0A1B6BXI8 A0A1A8QQS1 W5KCM7 A0A1A8DXA2 A0A1A8FHY1 A0A1A8BZ40 A0A1S3KH68 A0A3Q1I1Z5 A0A1A8GX15 A0A3P8XWD8 A0A1A8KEN3 A0A1A8QZ63 Q6ZM97 Q566T2 A0A1W4ZUI4 A0A210QT88 A0A3B4CED4 A0A3B4WNL1 A0A1A8ADB6 A0A1S3HMK8 A0A3B3T263 A0A1B6D306 A0A1L8HB15 A0A3P8XW97 A0A3Q3NAA3 A0A1B6M1M6 A0A3B4VPT0 A0A3B5A2Y4 F7A881 A0A2U9B6Y2 Q5FVB1 A4IHH8 A0A2J8V0Q0 W5UBQ7 A0A3Q1FP80 L8HVM2 Q9BVK2-2 A0A2I2ZXK9 A0A2K6C3K5 A0A2K5TXY9 A0A2D0SYZ3 A0A2J8MBN0 A0A2K5J2Y7 A0A2K5P155 A0A2I3LFC5 A0A067RF74 A0A2K6QEG2 M3ZBQ1 A0A3Q0TH41 F1MV61 A0A3P8TYA6 A0A1S3RG10 Q0P5D9 A0A3B3DQC3 A0A3P8ZTD2 A0A0F8APD9 A0A2J8V0P2 A0A2K6C3F4 K7ARD9 Q6NRA2 G2HE43 A0A024R5K5 Q9BVK2 H2Q4H1 F6WKY0 G3R7I9 A0A2K6C3I2 G7PNB7 H9EVM6 A0A2R9ABB6 A0A2K5TXV6 A0A3Q7T9G9 K9KBB2 F6VZR9 A0A2I4CUA4 A0A2K5J2R6 A0A3Q3DS33 A0A096MVL3 A0A2K5P150 A0A2Y9KDL1 U6DG13 W5P9G0 A0A2I3HB56 A0A3Q3JGS2 A0A2K6QEE8 M3WZ59 I3L8Z7 A0A0D9QUY0 A0A3P9IC15 A0A2I3T0N0
Ontologies
PATHWAY
GO
PANTHER
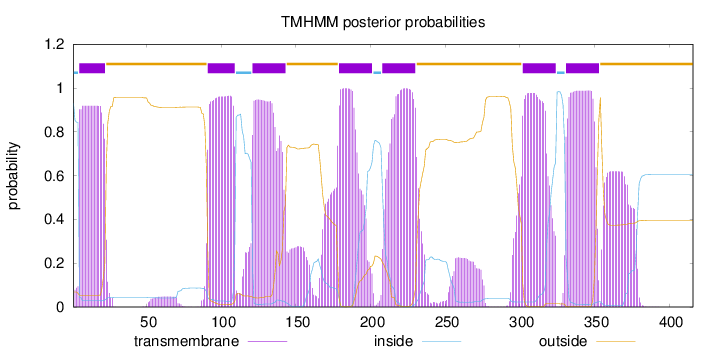
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
416
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
167.04937
Exp number, first 60 AAs:
17.46333
Total prob of N-in:
0.93138
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 22
outside
23 - 90
TMhelix
91 - 109
inside
110 - 120
TMhelix
121 - 143
outside
144 - 178
TMhelix
179 - 201
inside
202 - 207
TMhelix
208 - 230
outside
231 - 301
TMhelix
302 - 324
inside
325 - 330
TMhelix
331 - 353
outside
354 - 416
Population Genetic Test Statistics
Pi
190.539476
Theta
207.258602
Tajima's D
-1.073232
CLR
0.688547
CSRT
0.12284385780711
Interpretation
Uncertain
