Pre Gene Modal
BGIBMGA001209
Annotation
PREDICTED:_COP9_signalosome_complex_subunit_3_[Papilio_machaon]
Full name
COP9 signalosome complex subunit 3
Alternative Name
JAB1-containing signalosome subunit 3
Location in the cell
Cytoplasmic Reliability : 1.61 Extracellular Reliability : 1.275
Sequence
CDS
ATGGCTTCTCCTTTAGAACAATTTGTCAACAATGTGAGGACAATGTCGGCATCAGGTAATTTTCGCGATTTATATGAGATTATTGCAAAATCAGATGAAGTGTTGCAGAGGAATAGCTTCCACCTCAATACAGTACTTGAAACTCTTGATATCCAACAGCATTCATTGGGTGTTTTAGCAGTCCTGGTGGCCAAATTTTCCTTGCCTCCGGGCAATCCAGAAGTTGATAGGTCAACTATGTATCAACAGTTTCATGATTTTATCAATAACTGCAATGGTGAACAAGTTCGATTTGCAACTGATTTATATGCAGATTTGTGTCATCTACTCACAAATCATTTGGTAGAAATCAAACAACCAATAAGGGGCCTGGAAATATTAAAAAAGGCTATAAGAAAGATACAATTGTTTGATTCACAGCTGACCTCAATTCATGCTGATCTTTGCCAGCTCTGTTTGCTCTCAAAATGTATGAAGCCAGCTCTTGAGTTTTTGGACACTGACGTCACTGGGATTGGCAATGAGCTTGGCGGTATTAATGATTCCAAACATTTTCTACTATATTATTACTATGGGGGCATGATTTATACAGCAATGAAAAACTATGATAGAGCTTTGTATTTCTTTGAAGTTGTAGTGACTGTTCCGGCAATGGTTGTATCCCACATCATGTTGGAAGCATACAAGAAATATATTTTAGTGTCTCTTATACTTCATGGAAAGGTTTTGCCAATGCCGAAATACACATCTCAAGTGGTTTGTCGGTTTTTGAAACCGTTGTCAGCGCCATATCATGATTTAGTGGCATCCCAACATGCTGCAAGCAAACACCGCGAGATATTTGTCAGAGACAAGAATATGGGACTAGTTAATCAGGTTTTAAATTCAATGTATAAGAAGAATATTCAGAGATTGACAAAAACATTTTTAACGCTTTCGTTGAGCGATGTCGCTTCTCGCGTGGAATTATCTGGGCCTCTTCAAGCGGAAACATACATACTTAATATGATTGAAGAAGGCGAAATATATGCAATGATTAACCAAAAAGATGGTATGGTGGTGTTTCTAGATAGTCCAGAAAAATATGCATCACCGGAAACATTGTGTGTCTTAGAACAGCAAATGGCTGCTTGTACTAAACTTCACCAGTACATACAGGAAATGGATGTTCAGATACAAGTCAATCCACAGTATGTGAAGAAATCAGTTGGAAGTCACGATGAAGATATGCCCGGCACAAGTCAAAATACGAAGACAACATACACAATGTAA
Protein
MASPLEQFVNNVRTMSASGNFRDLYEIIAKSDEVLQRNSFHLNTVLETLDIQQHSLGVLAVLVAKFSLPPGNPEVDRSTMYQQFHDFINNCNGEQVRFATDLYADLCHLLTNHLVEIKQPIRGLEILKKAIRKIQLFDSQLTSIHADLCQLCLLSKCMKPALEFLDTDVTGIGNELGGINDSKHFLLYYYYGGMIYTAMKNYDRALYFFEVVVTVPAMVVSHIMLEAYKKYILVSLILHGKVLPMPKYTSQVVCRFLKPLSAPYHDLVASQHAASKHREIFVRDKNMGLVNQVLNSMYKKNIQRLTKTFLTLSLSDVASRVELSGPLQAETYILNMIEEGEIYAMINQKDGMVVFLDSPEKYASPETLCVLEQQMAACTKLHQYIQEMDVQIQVNPQYVKKSVGSHDEDMPGTSQNTKTTYTM
Summary
Description
Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of E3 ligase complexes, leading to modify the Ubl ligase activity (By similarity).
Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of SCF-type E3 ligase complexes, leading to decrease the Ubl ligase activity of SCF-type complexes such as SCF, CSA or DDB2. The complex is also involved in phosphorylation of p53/TP53, c-jun/JUN, IkappaBalpha/NFKBIA, ITPK1 and IRF8/ICSBP, possibly via its association with CK2 and PKD kinases. CSN-dependent phosphorylation of TP53 and JUN promotes and protects degradation by the Ubl system, respectively. Essential to maintain the survival of epiblast cells and thus the development of the postimplantation embryo.
Component of the COP9 signalosome complex (CSN), a complex involved in various cellular and developmental processes. The CSN complex is an essential regulator of the ubiquitin (Ubl) conjugation pathway by mediating the deneddylation of the cullin subunits of SCF-type E3 ligase complexes, leading to decrease the Ubl ligase activity of SCF-type complexes such as SCF, CSA or DDB2. The complex is also involved in phosphorylation of p53/TP53, c-jun/JUN, IkappaBalpha/NFKBIA, ITPK1 and IRF8/ICSBP, possibly via its association with CK2 and PKD kinases. CSN-dependent phosphorylation of TP53 and JUN promotes and protects degradation by the Ubl system, respectively. Essential to maintain the survival of epiblast cells and thus the development of the postimplantation embryo.
Subunit
Component of the CSN complex, probably composed of cops1, cops2, cops3, cops4, cops5, cops6, cops7, cops8 and cops9.
Component of the CSN complex, composed of COPS1/GPS1, COPS2, COPS3, COPS4, COPS5, COPS6, COPS7 (COPS7A or COPS7B), COPS8 and COPS9 (PubMed:9707402). In the complex, it probably interacts directly with COPS1, COPS4, COPS8 and COPS9. Interacts with CK2 and PKD. Interacts with the translation initiation factor EIF3S6 and IKBKG (By similarity).
Component of the CSN complex, composed of COPS1/GPS1, COPS2, COPS3, COPS4, COPS5, COPS6, COPS7 (COPS7A or COPS7B), COPS8 and COPS9 (PubMed:9707402). In the complex, it probably interacts directly with COPS1, COPS4, COPS8 and COPS9. Interacts with CK2 and PKD. Interacts with the translation initiation factor EIF3S6 and IKBKG (By similarity).
Similarity
Belongs to the CSN3 family.
Keywords
Complete proteome
Cytoplasm
Nucleus
Reference proteome
Signalosome
Acetylation
Phosphoprotein
Feature
chain COP9 signalosome complex subunit 3
Uniprot
H9IVC9
A0A3S2NEW8
S4P2Q8
A0A2A4JUC9
A0A2H1VGU3
A0A194RHJ8
+ More
A0A212ELY1 A0A194QEV3 A0A2A4JUK7 E2BPK5 A0A151X550 A0A195DQE1 A0A195EVE0 F4X259 A0A158NAJ0 A0A026WXI7 A0A0L7QN33 A0A1W4XU92 A0A067QS27 A0A2J7RAV1 A0A195CK47 A0A2A3ESB6 C3YWE6 A0A2R5LBI0 A0A1S3KEP4 A0A310SE83 A0A131XNX0 E0VSQ7 A0A1B6IIE3 K1QYH6 A0A154PNP6 A0A087UWZ5 A0A1B6H1Y0 M3ZJW5 G3NPQ9 W5M1F6 H3ANJ4 A0A3Q2ZTV0 A0A2R7WLN2 Q6P2U9 A0A3Q2E8K3 W5KBN9 B2GTE8 A0A0S7KKQ0 A0A3Q3IFH9 A0A3B3XQH8 A0A087YJ13 A0A3B3TL63 A0A3P9QD43 A0A3B4CUZ0 A0A3Q2PPM2 A0A3Q2X869 A0A3Q0SNM9 I3KUD0 A0A3P8QPA8 A0A3Q3KGS2 A0A146KXE6 A0A3P8WYG2 A0A3B3BU21 A0A3P8TWL2 A0A2D0SAL5 A0A2I4CBH4 A0A3B4YGE1 A0A3Q1GAV8 A0A3B4ZRR5 A0A3Q1H0L1 A0A3Q1CVU4 A0A3B4U783 A0A1W4ZB82 A0A060X8M2 B5X4W9 A0A1A7YZ90 A0A1Y1K187 A0A131X9K8 A0A0K8RCJ8 A0A2U9CL19 C1BKN9 A0A3B3QWV8 A0A3P8XLB8 A0A1B6L0M0 A0A060XH69 A0A1A8DII7 A0A1A8MR52 A0A3P9HM57 L7M8S2 A0A1A8G9P5 A0A224YQL2 A0A131Z2A0 Q3UQL2 O88543 A0A1A8CIZ2 A0A1A8HVR1 A0A1A8K9G8 A0A1A8GS95 A0A1A8RRL4 E2RD70 A0A3Q7TJE0 M3X663 A0A1A8ACH3 A0A1A8UPF1
A0A212ELY1 A0A194QEV3 A0A2A4JUK7 E2BPK5 A0A151X550 A0A195DQE1 A0A195EVE0 F4X259 A0A158NAJ0 A0A026WXI7 A0A0L7QN33 A0A1W4XU92 A0A067QS27 A0A2J7RAV1 A0A195CK47 A0A2A3ESB6 C3YWE6 A0A2R5LBI0 A0A1S3KEP4 A0A310SE83 A0A131XNX0 E0VSQ7 A0A1B6IIE3 K1QYH6 A0A154PNP6 A0A087UWZ5 A0A1B6H1Y0 M3ZJW5 G3NPQ9 W5M1F6 H3ANJ4 A0A3Q2ZTV0 A0A2R7WLN2 Q6P2U9 A0A3Q2E8K3 W5KBN9 B2GTE8 A0A0S7KKQ0 A0A3Q3IFH9 A0A3B3XQH8 A0A087YJ13 A0A3B3TL63 A0A3P9QD43 A0A3B4CUZ0 A0A3Q2PPM2 A0A3Q2X869 A0A3Q0SNM9 I3KUD0 A0A3P8QPA8 A0A3Q3KGS2 A0A146KXE6 A0A3P8WYG2 A0A3B3BU21 A0A3P8TWL2 A0A2D0SAL5 A0A2I4CBH4 A0A3B4YGE1 A0A3Q1GAV8 A0A3B4ZRR5 A0A3Q1H0L1 A0A3Q1CVU4 A0A3B4U783 A0A1W4ZB82 A0A060X8M2 B5X4W9 A0A1A7YZ90 A0A1Y1K187 A0A131X9K8 A0A0K8RCJ8 A0A2U9CL19 C1BKN9 A0A3B3QWV8 A0A3P8XLB8 A0A1B6L0M0 A0A060XH69 A0A1A8DII7 A0A1A8MR52 A0A3P9HM57 L7M8S2 A0A1A8G9P5 A0A224YQL2 A0A131Z2A0 Q3UQL2 O88543 A0A1A8CIZ2 A0A1A8HVR1 A0A1A8K9G8 A0A1A8GS95 A0A1A8RRL4 E2RD70 A0A3Q7TJE0 M3X663 A0A1A8ACH3 A0A1A8UPF1
Pubmed
19121390
23622113
26354079
22118469
20798317
21719571
+ More
21347285 24508170 30249741 24845553 18563158 20566863 22992520 23542700 9215903 25329095 25186727 26823975 24487278 29451363 24755649 20433749 28004739 28049606 29240929 25069045 17554307 25576852 28797301 26830274 10349636 11042159 11076861 11217851 12466851 16141073 21183079 9707402 15489334 12972600 16341006 17975172
21347285 24508170 30249741 24845553 18563158 20566863 22992520 23542700 9215903 25329095 25186727 26823975 24487278 29451363 24755649 20433749 28004739 28049606 29240929 25069045 17554307 25576852 28797301 26830274 10349636 11042159 11076861 11217851 12466851 16141073 21183079 9707402 15489334 12972600 16341006 17975172
EMBL
BABH01000896
RSAL01000152
RVE45712.1
GAIX01009887
JAA82673.1
NWSH01000642
+ More
PCG75083.1 ODYU01002495 SOQ40046.1 KQ460205 KPJ16914.1 AGBW02013971 OWR42486.1 KQ459299 KPJ01996.1 PCG75082.1 GL449633 EFN82409.1 KQ982519 KYQ55545.1 KQ980612 KYN15125.1 KQ981954 KYN32230.1 GL888565 EGI59475.1 ADTU01010338 KK107077 QOIP01000010 EZA60538.1 RLU17349.1 KQ414860 KOC60062.1 KK852999 KDR12669.1 NEVH01006564 PNF37957.1 KQ977642 KYN01096.1 KZ288193 PBC34019.1 GG666561 EEN55190.1 GGLE01002758 MBY06884.1 KQ770827 OAD52585.1 GEFM01006672 JAP69124.1 AAZO01005171 DS235755 EEB16413.1 GECU01025059 GECU01021014 JAS82647.1 JAS86692.1 JH818444 EKC42032.1 KQ435007 KZC13479.1 KK122078 KFM81884.1 GECZ01001121 JAS68648.1 AHAT01031987 AFYH01167875 AFYH01167876 AFYH01167877 KK855050 PTY20523.1 BC045415 BC064288 BC165863 AAI65863.1 GBYX01208248 JAO77163.1 AYCK01000799 AERX01002154 GDHC01019209 JAP99419.1 FR904885 CDQ73180.1 BT046088 ACI34350.1 HADW01001214 HADX01013209 SBP35441.1 GEZM01097366 JAV54248.1 GEFH01004938 JAP63643.1 GADI01005007 JAA68801.1 CP026260 AWP17304.1 BT075168 ACO09592.1 GEBQ01022767 JAT17210.1 FR905370 CDQ78776.1 HAEA01005387 SBQ33867.1 HAEF01017780 HAEG01015354 SBR58939.1 GACK01004772 JAA60262.1 HAEB01021266 SBQ67793.1 GFPF01006903 MAA18049.1 GEDV01003499 JAP85058.1 AK142318 BAE25027.1 AF071313 BC068179 HADZ01014790 SBP78731.1 HAED01002635 SBQ88483.1 HAEE01008933 SBR28983.1 HAEC01005769 SBQ73846.1 HAEI01009758 SBS08676.1 AAEX03003732 AANG04000740 HADY01013450 SBP51935.1 HAEJ01009029 SBS49486.1
PCG75083.1 ODYU01002495 SOQ40046.1 KQ460205 KPJ16914.1 AGBW02013971 OWR42486.1 KQ459299 KPJ01996.1 PCG75082.1 GL449633 EFN82409.1 KQ982519 KYQ55545.1 KQ980612 KYN15125.1 KQ981954 KYN32230.1 GL888565 EGI59475.1 ADTU01010338 KK107077 QOIP01000010 EZA60538.1 RLU17349.1 KQ414860 KOC60062.1 KK852999 KDR12669.1 NEVH01006564 PNF37957.1 KQ977642 KYN01096.1 KZ288193 PBC34019.1 GG666561 EEN55190.1 GGLE01002758 MBY06884.1 KQ770827 OAD52585.1 GEFM01006672 JAP69124.1 AAZO01005171 DS235755 EEB16413.1 GECU01025059 GECU01021014 JAS82647.1 JAS86692.1 JH818444 EKC42032.1 KQ435007 KZC13479.1 KK122078 KFM81884.1 GECZ01001121 JAS68648.1 AHAT01031987 AFYH01167875 AFYH01167876 AFYH01167877 KK855050 PTY20523.1 BC045415 BC064288 BC165863 AAI65863.1 GBYX01208248 JAO77163.1 AYCK01000799 AERX01002154 GDHC01019209 JAP99419.1 FR904885 CDQ73180.1 BT046088 ACI34350.1 HADW01001214 HADX01013209 SBP35441.1 GEZM01097366 JAV54248.1 GEFH01004938 JAP63643.1 GADI01005007 JAA68801.1 CP026260 AWP17304.1 BT075168 ACO09592.1 GEBQ01022767 JAT17210.1 FR905370 CDQ78776.1 HAEA01005387 SBQ33867.1 HAEF01017780 HAEG01015354 SBR58939.1 GACK01004772 JAA60262.1 HAEB01021266 SBQ67793.1 GFPF01006903 MAA18049.1 GEDV01003499 JAP85058.1 AK142318 BAE25027.1 AF071313 BC068179 HADZ01014790 SBP78731.1 HAED01002635 SBQ88483.1 HAEE01008933 SBR28983.1 HAEC01005769 SBQ73846.1 HAEI01009758 SBS08676.1 AAEX03003732 AANG04000740 HADY01013450 SBP51935.1 HAEJ01009029 SBS49486.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000008237 UP000075809 UP000078492 UP000078541 UP000007755 UP000005205 UP000053097 UP000279307 UP000053825 UP000192223 UP000027135 UP000235965 UP000078542 UP000242457 UP000001554 UP000085678 UP000009046 UP000005408 UP000076502 UP000054359 UP000002852 UP000007635 UP000018468 UP000008672 UP000264800 UP000000437 UP000265020 UP000018467 UP000261600 UP000261480 UP000028760 UP000261500 UP000242638 UP000261440 UP000265000 UP000264840 UP000261340 UP000005207 UP000265100 UP000261640 UP000265120 UP000261560 UP000265080 UP000221080 UP000192220 UP000261360 UP000257200 UP000261400 UP000265040 UP000257160 UP000261420 UP000192224 UP000193380 UP000087266 UP000246464 UP000261540 UP000265140 UP000265200 UP000000589 UP000002254 UP000286640 UP000011712
UP000008237 UP000075809 UP000078492 UP000078541 UP000007755 UP000005205 UP000053097 UP000279307 UP000053825 UP000192223 UP000027135 UP000235965 UP000078542 UP000242457 UP000001554 UP000085678 UP000009046 UP000005408 UP000076502 UP000054359 UP000002852 UP000007635 UP000018468 UP000008672 UP000264800 UP000000437 UP000265020 UP000018467 UP000261600 UP000261480 UP000028760 UP000261500 UP000242638 UP000261440 UP000265000 UP000264840 UP000261340 UP000005207 UP000265100 UP000261640 UP000265120 UP000261560 UP000265080 UP000221080 UP000192220 UP000261360 UP000257200 UP000261400 UP000265040 UP000257160 UP000261420 UP000192224 UP000193380 UP000087266 UP000246464 UP000261540 UP000265140 UP000265200 UP000000589 UP000002254 UP000286640 UP000011712
Pfam
PF01399 PCI
Interpro
Gene 3D
ProteinModelPortal
H9IVC9
A0A3S2NEW8
S4P2Q8
A0A2A4JUC9
A0A2H1VGU3
A0A194RHJ8
+ More
A0A212ELY1 A0A194QEV3 A0A2A4JUK7 E2BPK5 A0A151X550 A0A195DQE1 A0A195EVE0 F4X259 A0A158NAJ0 A0A026WXI7 A0A0L7QN33 A0A1W4XU92 A0A067QS27 A0A2J7RAV1 A0A195CK47 A0A2A3ESB6 C3YWE6 A0A2R5LBI0 A0A1S3KEP4 A0A310SE83 A0A131XNX0 E0VSQ7 A0A1B6IIE3 K1QYH6 A0A154PNP6 A0A087UWZ5 A0A1B6H1Y0 M3ZJW5 G3NPQ9 W5M1F6 H3ANJ4 A0A3Q2ZTV0 A0A2R7WLN2 Q6P2U9 A0A3Q2E8K3 W5KBN9 B2GTE8 A0A0S7KKQ0 A0A3Q3IFH9 A0A3B3XQH8 A0A087YJ13 A0A3B3TL63 A0A3P9QD43 A0A3B4CUZ0 A0A3Q2PPM2 A0A3Q2X869 A0A3Q0SNM9 I3KUD0 A0A3P8QPA8 A0A3Q3KGS2 A0A146KXE6 A0A3P8WYG2 A0A3B3BU21 A0A3P8TWL2 A0A2D0SAL5 A0A2I4CBH4 A0A3B4YGE1 A0A3Q1GAV8 A0A3B4ZRR5 A0A3Q1H0L1 A0A3Q1CVU4 A0A3B4U783 A0A1W4ZB82 A0A060X8M2 B5X4W9 A0A1A7YZ90 A0A1Y1K187 A0A131X9K8 A0A0K8RCJ8 A0A2U9CL19 C1BKN9 A0A3B3QWV8 A0A3P8XLB8 A0A1B6L0M0 A0A060XH69 A0A1A8DII7 A0A1A8MR52 A0A3P9HM57 L7M8S2 A0A1A8G9P5 A0A224YQL2 A0A131Z2A0 Q3UQL2 O88543 A0A1A8CIZ2 A0A1A8HVR1 A0A1A8K9G8 A0A1A8GS95 A0A1A8RRL4 E2RD70 A0A3Q7TJE0 M3X663 A0A1A8ACH3 A0A1A8UPF1
A0A212ELY1 A0A194QEV3 A0A2A4JUK7 E2BPK5 A0A151X550 A0A195DQE1 A0A195EVE0 F4X259 A0A158NAJ0 A0A026WXI7 A0A0L7QN33 A0A1W4XU92 A0A067QS27 A0A2J7RAV1 A0A195CK47 A0A2A3ESB6 C3YWE6 A0A2R5LBI0 A0A1S3KEP4 A0A310SE83 A0A131XNX0 E0VSQ7 A0A1B6IIE3 K1QYH6 A0A154PNP6 A0A087UWZ5 A0A1B6H1Y0 M3ZJW5 G3NPQ9 W5M1F6 H3ANJ4 A0A3Q2ZTV0 A0A2R7WLN2 Q6P2U9 A0A3Q2E8K3 W5KBN9 B2GTE8 A0A0S7KKQ0 A0A3Q3IFH9 A0A3B3XQH8 A0A087YJ13 A0A3B3TL63 A0A3P9QD43 A0A3B4CUZ0 A0A3Q2PPM2 A0A3Q2X869 A0A3Q0SNM9 I3KUD0 A0A3P8QPA8 A0A3Q3KGS2 A0A146KXE6 A0A3P8WYG2 A0A3B3BU21 A0A3P8TWL2 A0A2D0SAL5 A0A2I4CBH4 A0A3B4YGE1 A0A3Q1GAV8 A0A3B4ZRR5 A0A3Q1H0L1 A0A3Q1CVU4 A0A3B4U783 A0A1W4ZB82 A0A060X8M2 B5X4W9 A0A1A7YZ90 A0A1Y1K187 A0A131X9K8 A0A0K8RCJ8 A0A2U9CL19 C1BKN9 A0A3B3QWV8 A0A3P8XLB8 A0A1B6L0M0 A0A060XH69 A0A1A8DII7 A0A1A8MR52 A0A3P9HM57 L7M8S2 A0A1A8G9P5 A0A224YQL2 A0A131Z2A0 Q3UQL2 O88543 A0A1A8CIZ2 A0A1A8HVR1 A0A1A8K9G8 A0A1A8GS95 A0A1A8RRL4 E2RD70 A0A3Q7TJE0 M3X663 A0A1A8ACH3 A0A1A8UPF1
PDB
4WSN
E-value=1.92375e-138,
Score=1262
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
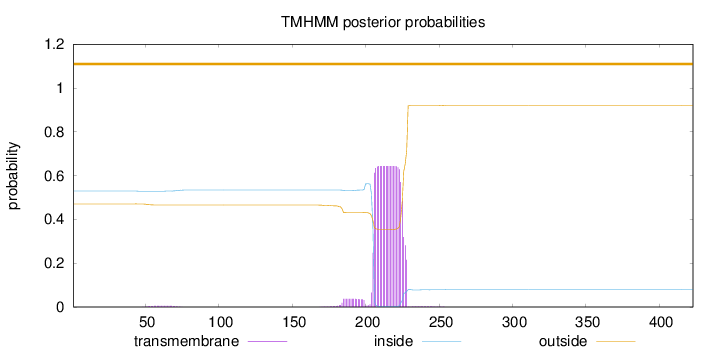
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.51395
Exp number, first 60 AAs:
0.04591
Total prob of N-in:
0.52922
outside
1 - 423
Population Genetic Test Statistics
Pi
169.288052
Theta
132.20133
Tajima's D
0.892649
CLR
0.00399
CSRT
0.628168591570421
Interpretation
Uncertain
