Gene
KWMTBOMO07843
Pre Gene Modal
BGIBMGA001211
Annotation
PREDICTED:_sterol_regulatory_element-binding_protein_2_[Amyelois_transitella]
Transcription factor
Location in the cell
Nuclear Reliability : 3.025
Sequence
CDS
ATGGATATTGATCCTTTTATGAATGCGGACGTTTTCAATGTCCACGACATTGCAGAAATTGAAGATTTTTTAAATAATTGCGATGGGGATTTTATGACAAAATTAGAACAGGAACTTACTTTTGTGGACAATACAAATGAAACAGGGCTGTTGGACATTGATACGAAATGCAAGCCTGAAATTTCATCATCTCCACAAGTTTCGCCGAATTATAATCATACCGGGAATCCAGTGATGTCACAGCACACGCATAGAATTAGGCCAACGCCCCCGATCAGTAAAACAATACAAGGTGGATATATTAAAGAAGAGAAATTCAATTTGCCAACATCCCTTGAGAATGCGTCCGTGACGTTTCCTGAAGTTATACCACAACAGAGGACTCAAGTTCAAAATCAGCCGATGATTTTTCAACAAGTAGTACAGAGCCCCGTGTTTGTTAATTTGGGTCCGGCTGGAAATATCCAACCTATGCCTGACAAAAGGTCAAATGTCATTCAAGTTGATAATGTTCCACAAGTAAATAAAACTAGCCCCCTAAAACAGCAGAGTCAACCTCTACTGATACAAAACAATCCCAAAGGTGTACCTTTTATATTAAAAAGTTCTGATTCTAATTTCTCACCAGTGTTATTGCAGTCGAATATCATTAACCCAAAGACACAAACGTTGATGTATACCAGTACTCCATTACAGGTTTCGAGCACAAACCAAAATATTATTACAAATGCAAAGGCAGGATCGGAGACAACACCCATACACACATTTTTTACAAACAGTAACGGTACTACTTTATTGACGGGAATACCTGCAGTTGTACTAGAAGGTGATAAAATTGCTTTAAACCCTGCACCAAACATTGGACCTCCAAAAGTGAAAGAAGTGAAACGCAGTGCTCACAATGCCATTGAAAGACGTTACAGAACTAGTATTAATGATAGAATAGTGGAATTGAAGAATATGCTTGTTGGTGAAGAGGCCAAGTTGAACAAGTCAGCAATATTGCGTAAAACAATTGAGTACATTAAGTATTTGCGAAGTCAGAACACTAGGTTAAAACAAGAAAATATAGCTCTTAAACAGGTTTGTCAAAAGTCTGGAGTTAAAGAGCCAATATTTGAAGGAGGTTATACTCCACCACACAGTGATGTGTCATCACCTTATCATTCTCCACATTCAATTGACAGCGACACACCTTCTTCACCAGAATACAAGACTGAAGATAAATACTCAAAGATAGTAATGGGTATGGGTGATCATTCACGACTGGCGTTATGTGCCTTCATGGTTGGGCTTATTGCTTTCAACCCATTTAGCACATTCTTTAGCAGTTTTACAACAGAGTATACAACTACAGATTTCAATGCAAGAATTGATCAAAGAAAAATTCTATCAGAAGATGATTCAAGTTATGAAGGATCATCTTGGGCGATTTGCGTTTTTAATACATTCTTTATATATCTTGTTAACTTTGTAATACTCGGAGGCTGTTTGGTAAAGTTACTAGTTTATGGAGATTCTGTACCAAAAAGTCAGTCCAAGGAAGCTGGTTTATTCTACAAACATAAAAGGCTTGCTGGCAACCATCTTAAAAAGGGCGATCTAAATAATACGCGACTAGAACTCCATCGAGCATTGGAAGCATGTGGAAGAAGTGTCCAGGCAGGTGGCCCAGGTCAAGCTAAGTACACGGCCCTCACAGCGGCTGTGCTTCGGCAGTTGCTGCAGAGATTACCGTTCGGTGGCTTCCTTGCACGACGCGCCGGAGATCTGTGGAGGGACAGTCCAACAAGACGAGCTACGTTGCACTGGGCAACGGAGGTTTCTTCTGTTTCGCATCAACTAGCCCAGCTGGAGATCCTGTCGGATCACAGAGGGCGCAGCGAACGATTGTTGCTAGCGCTCCAAGCGATTAATTTGGCTGAGGTCGCCGGGAACAGACAATTACTGGCCGACACGTACGTCACGGCGGCTTTAGTCTTTAAAGATTATATGCCTAAGATCGGAAACTGGTTGTGCGGGTAA
Protein
MDIDPFMNADVFNVHDIAEIEDFLNNCDGDFMTKLEQELTFVDNTNETGLLDIDTKCKPEISSSPQVSPNYNHTGNPVMSQHTHRIRPTPPISKTIQGGYIKEEKFNLPTSLENASVTFPEVIPQQRTQVQNQPMIFQQVVQSPVFVNLGPAGNIQPMPDKRSNVIQVDNVPQVNKTSPLKQQSQPLLIQNNPKGVPFILKSSDSNFSPVLLQSNIINPKTQTLMYTSTPLQVSSTNQNIITNAKAGSETTPIHTFFTNSNGTTLLTGIPAVVLEGDKIALNPAPNIGPPKVKEVKRSAHNAIERRYRTSINDRIVELKNMLVGEEAKLNKSAILRKTIEYIKYLRSQNTRLKQENIALKQVCQKSGVKEPIFEGGYTPPHSDVSSPYHSPHSIDSDTPSSPEYKTEDKYSKIVMGMGDHSRLALCAFMVGLIAFNPFSTFFSSFTTEYTTTDFNARIDQRKILSEDDSSYEGSSWAICVFNTFFIYLVNFVILGGCLVKLLVYGDSVPKSQSKEAGLFYKHKRLAGNHLKKGDLNNTRLELHRALEACGRSVQAGGPGQAKYTALTAAVLRQLLQRLPFGGFLARRAGDLWRDSPTRRATLHWATEVSSVSHQLAQLEILSDHRGRSERLLLALQAINLAEVAGNRQLLADTYVTAALVFKDYMPKIGNWLCG
Summary
Uniprot
H9IVD1
A0A2H1VIL4
A0A2A4JTI6
A0A212ELX8
A0A194Q9A4
A0A194RLQ1
+ More
A0A1E1WVK0 A0A1Y1MFB5 A0A067QQZ3 A0A1B6EGK4 A0A1W4WSR3 A0A0F7IKE5 D2A113 A0A2J7RQE5 A0A1J1IHR2 A0A0N0BGW5 A0A1B6H6H1 A0A1B6MLF2 F1RDU4 E2BFH0 A0A1B6G2C4 A0A0M4B676 A0A310SBQ8 E0VES1 A0A2D0PPZ9 A0A2D0PSM2 H2M0A8 A0A154NX90 A6XLD8 A0A3P9J025 A0A3P9MGE9 A0A0L7RK29 A0A146MVA7 A0A1U8D5I7 A0A182YCN2 U4U7N4 A0A026W4I3 A0A146QFT9 A0A1S4FQK8 A0A3Q3BNN1 A0A023EWQ7 A0A154MDH6 A0A151P4D4 A0A182QXP7 A0A0B8RP38 F6U0V4 A0A146TLU8 A0A3Q2Q7A3 N6U3G2 A0A2R3ZDF3 A0A3Q2Q6F8 A0A146W0U6 A0A182G9E1 A0A0A7KZK1 A0A146S9A0 A0A084VJ23 A0A3L8D502 A0A385MCM3 Q4S875 A0A2A3EGT0 V9IIG4 A0A3Q3NEP2 M9NGV6 A0A3B4TXF1 A0A3Q3NEN1 A0A3P8WHT0 A0A3B3C136 A0A3P8WBW5 A0A3B4ZZL6 A0A3B4TY15 A0A3Q3T2K7 A0A182RZ29 A0A1L1RPI1 A0A1W7RJ46 A0A226PTJ5 A0A3Q0GKX1 A0A3B4E3Z7 A0A3B4Z7I8 B0XGW8 R4GLV5 A0A3Q4GFF9 Q90ZM5 A0A2I4BRI0 A0A1L8EYS0 A0A151P412 A0A151P404 A0A151P3Y7 A0A182T7F5 J3SDG9 A0A3Q4MA31 A0A3B5A967
A0A1E1WVK0 A0A1Y1MFB5 A0A067QQZ3 A0A1B6EGK4 A0A1W4WSR3 A0A0F7IKE5 D2A113 A0A2J7RQE5 A0A1J1IHR2 A0A0N0BGW5 A0A1B6H6H1 A0A1B6MLF2 F1RDU4 E2BFH0 A0A1B6G2C4 A0A0M4B676 A0A310SBQ8 E0VES1 A0A2D0PPZ9 A0A2D0PSM2 H2M0A8 A0A154NX90 A6XLD8 A0A3P9J025 A0A3P9MGE9 A0A0L7RK29 A0A146MVA7 A0A1U8D5I7 A0A182YCN2 U4U7N4 A0A026W4I3 A0A146QFT9 A0A1S4FQK8 A0A3Q3BNN1 A0A023EWQ7 A0A154MDH6 A0A151P4D4 A0A182QXP7 A0A0B8RP38 F6U0V4 A0A146TLU8 A0A3Q2Q7A3 N6U3G2 A0A2R3ZDF3 A0A3Q2Q6F8 A0A146W0U6 A0A182G9E1 A0A0A7KZK1 A0A146S9A0 A0A084VJ23 A0A3L8D502 A0A385MCM3 Q4S875 A0A2A3EGT0 V9IIG4 A0A3Q3NEP2 M9NGV6 A0A3B4TXF1 A0A3Q3NEN1 A0A3P8WHT0 A0A3B3C136 A0A3P8WBW5 A0A3B4ZZL6 A0A3B4TY15 A0A3Q3T2K7 A0A182RZ29 A0A1L1RPI1 A0A1W7RJ46 A0A226PTJ5 A0A3Q0GKX1 A0A3B4E3Z7 A0A3B4Z7I8 B0XGW8 R4GLV5 A0A3Q4GFF9 Q90ZM5 A0A2I4BRI0 A0A1L8EYS0 A0A151P412 A0A151P404 A0A151P3Y7 A0A182T7F5 J3SDG9 A0A3Q4MA31 A0A3B5A967
Pubmed
EMBL
BABH01000899
BABH01000900
BABH01000901
BABH01000902
ODYU01002774
SOQ40679.1
+ More
NWSH01000642 PCG75086.1 AGBW02013971 OWR42483.1 KQ459299 KPJ01999.1 KQ460205 KPJ16911.1 GDQN01000052 JAT91002.1 GEZM01036620 JAV82566.1 KK853125 KDR11017.1 GEDC01000249 JAS37049.1 KP147920 AKG92757.1 KQ971338 EFA01599.1 NEVH01001340 PNF43047.1 CVRI01000053 CRK99799.1 KQ435771 KOX75141.1 GECU01037409 JAS70297.1 GEBQ01003233 JAT36744.1 FQ312026 GL448037 EFN85492.1 GECZ01013238 JAS56531.1 KR020013 ALB38958.1 KQ797156 OAD46986.1 DS235096 EEB11877.1 KQ434777 KZC04213.1 DQ836065 ABI33876.1 KQ414578 KOC71163.1 GCES01162754 JAQ23568.1 KB632144 ERL89082.1 KK107429 EZA50933.1 GCES01131263 JAQ55059.1 GAPW01000050 JAC13548.1 GCES01117546 GCES01063065 GCES01063064 JAR23259.1 AKHW03001118 KYO43789.1 AXCN02001227 GBSH01002607 JAG66419.1 AAMC01050816 AAMC01050817 AAMC01050818 GCES01092552 JAQ93770.1 APGK01041124 APGK01041125 KB740989 ENN76105.1 MG735210 AVR48442.1 GCES01063060 JAR23263.1 JXUM01050239 KQ561640 KXJ77916.1 KJ162572 AIZ50708.1 GCES01109194 JAQ77128.1 ATLV01013426 KE524855 KFB37967.1 QOIP01000014 RLU14928.1 MH633449 AYA57965.1 CAAE01014707 CAG03157.1 KZ288253 PBC30975.1 JR049085 AEY60855.1 JF502069 KU670831 AFH35105.1 ANN45572.1 AADN05000376 GDAY02000262 JAV51151.1 AWGT02000022 OXB82802.1 DS233070 EDS27855.1 AY029224 AAK50343.1 CM004482 OCT64473.1 KYO43788.1 KYO43786.1 KYO43787.1 JU176137 AFJ51660.1
NWSH01000642 PCG75086.1 AGBW02013971 OWR42483.1 KQ459299 KPJ01999.1 KQ460205 KPJ16911.1 GDQN01000052 JAT91002.1 GEZM01036620 JAV82566.1 KK853125 KDR11017.1 GEDC01000249 JAS37049.1 KP147920 AKG92757.1 KQ971338 EFA01599.1 NEVH01001340 PNF43047.1 CVRI01000053 CRK99799.1 KQ435771 KOX75141.1 GECU01037409 JAS70297.1 GEBQ01003233 JAT36744.1 FQ312026 GL448037 EFN85492.1 GECZ01013238 JAS56531.1 KR020013 ALB38958.1 KQ797156 OAD46986.1 DS235096 EEB11877.1 KQ434777 KZC04213.1 DQ836065 ABI33876.1 KQ414578 KOC71163.1 GCES01162754 JAQ23568.1 KB632144 ERL89082.1 KK107429 EZA50933.1 GCES01131263 JAQ55059.1 GAPW01000050 JAC13548.1 GCES01117546 GCES01063065 GCES01063064 JAR23259.1 AKHW03001118 KYO43789.1 AXCN02001227 GBSH01002607 JAG66419.1 AAMC01050816 AAMC01050817 AAMC01050818 GCES01092552 JAQ93770.1 APGK01041124 APGK01041125 KB740989 ENN76105.1 MG735210 AVR48442.1 GCES01063060 JAR23263.1 JXUM01050239 KQ561640 KXJ77916.1 KJ162572 AIZ50708.1 GCES01109194 JAQ77128.1 ATLV01013426 KE524855 KFB37967.1 QOIP01000014 RLU14928.1 MH633449 AYA57965.1 CAAE01014707 CAG03157.1 KZ288253 PBC30975.1 JR049085 AEY60855.1 JF502069 KU670831 AFH35105.1 ANN45572.1 AADN05000376 GDAY02000262 JAV51151.1 AWGT02000022 OXB82802.1 DS233070 EDS27855.1 AY029224 AAK50343.1 CM004482 OCT64473.1 KYO43788.1 KYO43786.1 KYO43787.1 JU176137 AFJ51660.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000027135
+ More
UP000192223 UP000007266 UP000235965 UP000183832 UP000053105 UP000000437 UP000008237 UP000009046 UP000221080 UP000001038 UP000076502 UP000265200 UP000265180 UP000053825 UP000189705 UP000076408 UP000030742 UP000053097 UP000264800 UP000050525 UP000075886 UP000008143 UP000265000 UP000019118 UP000069940 UP000249989 UP000030765 UP000279307 UP000242457 UP000261640 UP000261420 UP000265120 UP000261560 UP000261400 UP000075900 UP000000539 UP000198419 UP000261440 UP000261360 UP000002320 UP000261580 UP000192220 UP000186698 UP000075901
UP000192223 UP000007266 UP000235965 UP000183832 UP000053105 UP000000437 UP000008237 UP000009046 UP000221080 UP000001038 UP000076502 UP000265200 UP000265180 UP000053825 UP000189705 UP000076408 UP000030742 UP000053097 UP000264800 UP000050525 UP000075886 UP000008143 UP000265000 UP000019118 UP000069940 UP000249989 UP000030765 UP000279307 UP000242457 UP000261640 UP000261420 UP000265120 UP000261560 UP000261400 UP000075900 UP000000539 UP000198419 UP000261440 UP000261360 UP000002320 UP000261580 UP000192220 UP000186698 UP000075901
Interpro
SUPFAM
SSF47459
SSF47459
Gene 3D
ProteinModelPortal
H9IVD1
A0A2H1VIL4
A0A2A4JTI6
A0A212ELX8
A0A194Q9A4
A0A194RLQ1
+ More
A0A1E1WVK0 A0A1Y1MFB5 A0A067QQZ3 A0A1B6EGK4 A0A1W4WSR3 A0A0F7IKE5 D2A113 A0A2J7RQE5 A0A1J1IHR2 A0A0N0BGW5 A0A1B6H6H1 A0A1B6MLF2 F1RDU4 E2BFH0 A0A1B6G2C4 A0A0M4B676 A0A310SBQ8 E0VES1 A0A2D0PPZ9 A0A2D0PSM2 H2M0A8 A0A154NX90 A6XLD8 A0A3P9J025 A0A3P9MGE9 A0A0L7RK29 A0A146MVA7 A0A1U8D5I7 A0A182YCN2 U4U7N4 A0A026W4I3 A0A146QFT9 A0A1S4FQK8 A0A3Q3BNN1 A0A023EWQ7 A0A154MDH6 A0A151P4D4 A0A182QXP7 A0A0B8RP38 F6U0V4 A0A146TLU8 A0A3Q2Q7A3 N6U3G2 A0A2R3ZDF3 A0A3Q2Q6F8 A0A146W0U6 A0A182G9E1 A0A0A7KZK1 A0A146S9A0 A0A084VJ23 A0A3L8D502 A0A385MCM3 Q4S875 A0A2A3EGT0 V9IIG4 A0A3Q3NEP2 M9NGV6 A0A3B4TXF1 A0A3Q3NEN1 A0A3P8WHT0 A0A3B3C136 A0A3P8WBW5 A0A3B4ZZL6 A0A3B4TY15 A0A3Q3T2K7 A0A182RZ29 A0A1L1RPI1 A0A1W7RJ46 A0A226PTJ5 A0A3Q0GKX1 A0A3B4E3Z7 A0A3B4Z7I8 B0XGW8 R4GLV5 A0A3Q4GFF9 Q90ZM5 A0A2I4BRI0 A0A1L8EYS0 A0A151P412 A0A151P404 A0A151P3Y7 A0A182T7F5 J3SDG9 A0A3Q4MA31 A0A3B5A967
A0A1E1WVK0 A0A1Y1MFB5 A0A067QQZ3 A0A1B6EGK4 A0A1W4WSR3 A0A0F7IKE5 D2A113 A0A2J7RQE5 A0A1J1IHR2 A0A0N0BGW5 A0A1B6H6H1 A0A1B6MLF2 F1RDU4 E2BFH0 A0A1B6G2C4 A0A0M4B676 A0A310SBQ8 E0VES1 A0A2D0PPZ9 A0A2D0PSM2 H2M0A8 A0A154NX90 A6XLD8 A0A3P9J025 A0A3P9MGE9 A0A0L7RK29 A0A146MVA7 A0A1U8D5I7 A0A182YCN2 U4U7N4 A0A026W4I3 A0A146QFT9 A0A1S4FQK8 A0A3Q3BNN1 A0A023EWQ7 A0A154MDH6 A0A151P4D4 A0A182QXP7 A0A0B8RP38 F6U0V4 A0A146TLU8 A0A3Q2Q7A3 N6U3G2 A0A2R3ZDF3 A0A3Q2Q6F8 A0A146W0U6 A0A182G9E1 A0A0A7KZK1 A0A146S9A0 A0A084VJ23 A0A3L8D502 A0A385MCM3 Q4S875 A0A2A3EGT0 V9IIG4 A0A3Q3NEP2 M9NGV6 A0A3B4TXF1 A0A3Q3NEN1 A0A3P8WHT0 A0A3B3C136 A0A3P8WBW5 A0A3B4ZZL6 A0A3B4TY15 A0A3Q3T2K7 A0A182RZ29 A0A1L1RPI1 A0A1W7RJ46 A0A226PTJ5 A0A3Q0GKX1 A0A3B4E3Z7 A0A3B4Z7I8 B0XGW8 R4GLV5 A0A3Q4GFF9 Q90ZM5 A0A2I4BRI0 A0A1L8EYS0 A0A151P412 A0A151P404 A0A151P3Y7 A0A182T7F5 J3SDG9 A0A3Q4MA31 A0A3B5A967
PDB
1AM9
E-value=1.11398e-20,
Score=249
Ontologies
GO
GO:0016021
GO:0046983
GO:0006355
GO:1990798
GO:0031018
GO:0003323
GO:0001889
GO:0003896
GO:0003677
GO:0019901
GO:0005737
GO:0004879
GO:0005634
GO:0031065
GO:0032810
GO:0007623
GO:0009416
GO:0045444
GO:0003062
GO:0046676
GO:0008610
GO:0045542
GO:0042789
GO:1903146
GO:0019217
GO:0006351
GO:0000122
GO:0010867
GO:0008286
GO:0009749
GO:0016020
GO:0003682
GO:0045944
GO:0031647
GO:0009267
GO:1903214
GO:0009052
GO:0043565
GO:0005506
GO:0030145
GO:0050906
GO:0005515
GO:0016818
GO:0003707
GO:0051537
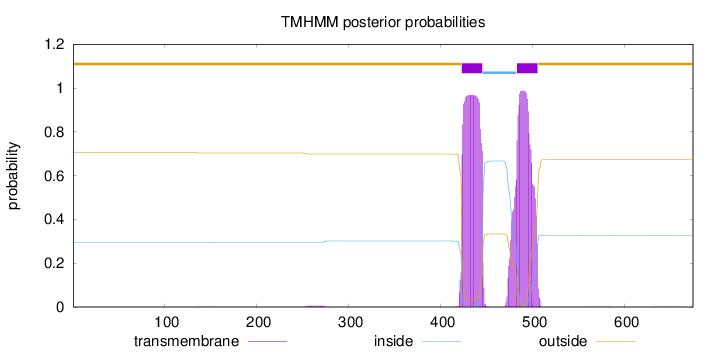
Topology
Length:
674
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.9670100000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.29579
outside
1 - 422
TMhelix
423 - 445
inside
446 - 482
TMhelix
483 - 505
outside
506 - 674
Population Genetic Test Statistics
Pi
275.104272
Theta
232.060315
Tajima's D
0.631979
CLR
0.000089
CSRT
0.556422178891055
Interpretation
Uncertain
