Gene
KWMTBOMO07841
Pre Gene Modal
BGIBMGA001212
Annotation
centrin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.447
Sequence
CDS
ATGGCAACCGCGGTCGCAACAATACAGAAAAAAACTGCAACTACCAACCCTGGACCAGGTGGTGTTCGTAAGAAATCTGGACCTAAATTTGAGCTAACCGAAGAACAAAAAAGAGACATCAAAGAAGCATTTGATTTATTTGATACAGAAAATACTGGAAAAATCGACACCAAAGAACTTAAAGTGGCAATTCGAGCCCTTGGTTTCGAACCAAAAAAAGAAGAAATAAAAAAAATGATTGCAGAGATAGACAAAGGTGATGGAAAAGTATCATTTGATGACTTTATGGAACTCATGTCAGTGAAGATGGCAGAAAAAGATACCAGGGAGGAAATCATGAAAGCATTTAAACTGTTTGATGATGACGAAACTGGTAAAATTTCATTCAAAAATTTGAAGAGAGTTGCAAAGGAACTTGGTGAAAATCTCACAGATGAAGAACTGCATGAAATGATAGATGAAGCTGATAGAGATGGGGATGGTGAAATAAATCAAGAGGAATTCTTGCGTATCATGAAAAAGACTAGTCTCTATTAA
Protein
MATAVATIQKKTATTNPGPGGVRKKSGPKFELTEEQKRDIKEAFDLFDTENTGKIDTKELKVAIRALGFEPKKEEIKKMIAEIDKGDGKVSFDDFMELMSVKMAEKDTREEIMKAFKLFDDDETGKISFKNLKRVAKELGENLTDEELHEMIDEADRDGDGEINQEEFLRIMKKTSLY
Summary
Uniprot
Q2F618
A0A2A4JTR9
A0A212ELW7
S4P7E0
A0A2H1VIK9
I4DKP8
+ More
A0A194RHV6 A0A194QAH8 A0A0L7KZW2 A0A2M3ZJZ4 A0A2M4C0I5 W5JGC2 A0A2M4AXW4 A0A182VUG3 B5M0Q9 A0A182PE83 A0A182F3J8 A0A2M4C0F2 H9IVD2 A0A182U6I1 A0A182QY57 A0A182K4V4 A0A182WW33 A0A182VMX0 A0A182L6D7 A0A182HGH8 A0A182MQ98 A0A182T686 A0A182YDR1 A0A182RIP8 A0A1S4H0J1 Q7PVJ3 A0A182NU43 A0A182IY95 A0A182H2A0 Q179F4 A0A0K8TKX8 A0A084WFZ7 U4UUI5 A0A1B0C1H0 A0A1Q3FEH8 B0WFF6 N6UKA8 D2A0V1 A0A1B0CN05 A0A1B0GA75 A0A1A9UPP5 A0A1A9ZP19 T1PK64 A0A1L8DM28 A0A1Y1JWA6 E0VM15 A0A067QNL7 W4ZKD9 A0A2J7PFI5 A0A087V299 G3WRI8 A0A1A9Y571 A0A2J7PFI0 A0A1W4W857 F6WWN8 A0A1J1HTY8 G1N1G4 A0A0A0A700 A0A093NCW4 A0A087QQR1 A0A093T696 A0A091X492 A0A1W4WIG3 F1NM71 B4JBM0 B3N008 K7FX63 G1SWQ3 A0A091JWD2 A0A093GYM0 A0A3B4YHY7 A0A3B4TWQ3 A0A3B0JYP5 A0A1B6CM56 J3S8D8 T1DCW4 T1GD07 A0A1B6LAR5 A0A0B8RZB0 A0A0L8GXM8 A0A091JAB0 H3CU91 A0A091IDW2 A0A098LZR9 A0A1W4W359 A0A094KU40 A0A091NJT5 A0A2D4ML54 T1G1D6 A0A2I4B175 A0A1W7RIH0 A0A3B4X2C1 A0A3B4TX62 A0A093H2L8
A0A194RHV6 A0A194QAH8 A0A0L7KZW2 A0A2M3ZJZ4 A0A2M4C0I5 W5JGC2 A0A2M4AXW4 A0A182VUG3 B5M0Q9 A0A182PE83 A0A182F3J8 A0A2M4C0F2 H9IVD2 A0A182U6I1 A0A182QY57 A0A182K4V4 A0A182WW33 A0A182VMX0 A0A182L6D7 A0A182HGH8 A0A182MQ98 A0A182T686 A0A182YDR1 A0A182RIP8 A0A1S4H0J1 Q7PVJ3 A0A182NU43 A0A182IY95 A0A182H2A0 Q179F4 A0A0K8TKX8 A0A084WFZ7 U4UUI5 A0A1B0C1H0 A0A1Q3FEH8 B0WFF6 N6UKA8 D2A0V1 A0A1B0CN05 A0A1B0GA75 A0A1A9UPP5 A0A1A9ZP19 T1PK64 A0A1L8DM28 A0A1Y1JWA6 E0VM15 A0A067QNL7 W4ZKD9 A0A2J7PFI5 A0A087V299 G3WRI8 A0A1A9Y571 A0A2J7PFI0 A0A1W4W857 F6WWN8 A0A1J1HTY8 G1N1G4 A0A0A0A700 A0A093NCW4 A0A087QQR1 A0A093T696 A0A091X492 A0A1W4WIG3 F1NM71 B4JBM0 B3N008 K7FX63 G1SWQ3 A0A091JWD2 A0A093GYM0 A0A3B4YHY7 A0A3B4TWQ3 A0A3B0JYP5 A0A1B6CM56 J3S8D8 T1DCW4 T1GD07 A0A1B6LAR5 A0A0B8RZB0 A0A0L8GXM8 A0A091JAB0 H3CU91 A0A091IDW2 A0A098LZR9 A0A1W4W359 A0A094KU40 A0A091NJT5 A0A2D4ML54 T1G1D6 A0A2I4B175 A0A1W7RIH0 A0A3B4X2C1 A0A3B4TX62 A0A093H2L8
Pubmed
22118469
23622113
22651552
26354079
26227816
20920257
+ More
23761445 19166301 19121390 20966253 25244985 12364791 26483478 17510324 26369729 24438588 23537049 18362917 19820115 25315136 28004739 20566863 24845553 21709235 17495919 20838655 15592404 17994087 17381049 21993624 23025625 23758969 25727380 25476704 15496914 23254933 26358130
23761445 19166301 19121390 20966253 25244985 12364791 26483478 17510324 26369729 24438588 23537049 18362917 19820115 25315136 28004739 20566863 24845553 21709235 17495919 20838655 15592404 17994087 17381049 21993624 23025625 23758969 25727380 25476704 15496914 23254933 26358130
EMBL
DQ311254
ABD36199.1
NWSH01000642
PCG75088.1
AGBW02013971
OWR42482.1
+ More
GAIX01006421 JAA86139.1 ODYU01002774 SOQ40680.1 AK401866 BAM18488.1 KQ460205 KPJ16910.1 KQ459299 KPJ02000.1 JTDY01004095 KOB68611.1 GGFM01008093 MBW28844.1 GGFJ01009643 MBW58784.1 ADMH02001595 ETN61930.1 GGFK01012318 MBW45639.1 EU930245 ACH56873.1 GGFJ01009642 MBW58783.1 BABH01000902 AXCN02000773 APCN01004235 AXCM01000998 AAAB01008984 EAA43434.4 JXUM01104930 KQ564927 KXJ71562.1 CH477353 EAT42852.1 GDAI01002790 JAI14813.1 ATLV01023415 KE525343 KFB49141.1 KB632374 ERL93831.1 JXJN01024045 GFDL01009089 JAV25956.1 DS231917 EDS26193.1 APGK01005693 APGK01005750 APGK01015356 APGK01015617 KB739627 KB739555 KB736406 KB736386 ENN82190.1 ENN82223.1 ENN83206.1 ENN83211.1 KQ971338 EFA02905.2 AJWK01019584 CCAG010019840 KA648353 AFP62982.1 GFDF01006670 JAV07414.1 GEZM01098974 JAV53599.1 DS235286 EEB14421.1 KK853132 KDR10884.1 AAGJ04046523 NEVH01025657 PNF15093.1 KL477171 KFO06741.1 AEFK01230660 AEFK01230661 PNF15091.1 CVRI01000020 CRK90836.1 KL870974 KGL89846.1 KL224695 KFW62758.1 KL225814 KFM03565.1 KL425250 KFW90109.1 KL411511 KFR08331.1 AADN05000020 CH916368 EDW02955.1 CH902638 EDV35374.1 AGCU01008173 AAGW02031682 KK531768 KFP28238.1 KL205624 KFV72230.1 OUUW01000010 SPP86193.1 GEDC01022833 JAS14465.1 JU174157 GBEX01000681 AFJ49683.1 JAI13879.1 GAAZ01000499 GBKC01000707 GBKD01000453 JAA97444.1 JAG45363.1 JAG47165.1 CAQQ02060992 GEBQ01029129 GEBQ01019242 JAT10848.1 JAT20735.1 GBSH01000642 JAG68383.1 KQ419989 KOF81806.1 KK501764 KFP17527.1 KL218474 KFP05688.1 GBSI01000528 JAC95968.1 KL354027 KFZ60847.1 KL384202 KFP89337.1 IACM01108630 IACM01108631 LAB33798.1 AMQM01002998 KB095959 ESO09582.1 GDAY02000469 JAV50944.1 KK381991 KFV48978.1
GAIX01006421 JAA86139.1 ODYU01002774 SOQ40680.1 AK401866 BAM18488.1 KQ460205 KPJ16910.1 KQ459299 KPJ02000.1 JTDY01004095 KOB68611.1 GGFM01008093 MBW28844.1 GGFJ01009643 MBW58784.1 ADMH02001595 ETN61930.1 GGFK01012318 MBW45639.1 EU930245 ACH56873.1 GGFJ01009642 MBW58783.1 BABH01000902 AXCN02000773 APCN01004235 AXCM01000998 AAAB01008984 EAA43434.4 JXUM01104930 KQ564927 KXJ71562.1 CH477353 EAT42852.1 GDAI01002790 JAI14813.1 ATLV01023415 KE525343 KFB49141.1 KB632374 ERL93831.1 JXJN01024045 GFDL01009089 JAV25956.1 DS231917 EDS26193.1 APGK01005693 APGK01005750 APGK01015356 APGK01015617 KB739627 KB739555 KB736406 KB736386 ENN82190.1 ENN82223.1 ENN83206.1 ENN83211.1 KQ971338 EFA02905.2 AJWK01019584 CCAG010019840 KA648353 AFP62982.1 GFDF01006670 JAV07414.1 GEZM01098974 JAV53599.1 DS235286 EEB14421.1 KK853132 KDR10884.1 AAGJ04046523 NEVH01025657 PNF15093.1 KL477171 KFO06741.1 AEFK01230660 AEFK01230661 PNF15091.1 CVRI01000020 CRK90836.1 KL870974 KGL89846.1 KL224695 KFW62758.1 KL225814 KFM03565.1 KL425250 KFW90109.1 KL411511 KFR08331.1 AADN05000020 CH916368 EDW02955.1 CH902638 EDV35374.1 AGCU01008173 AAGW02031682 KK531768 KFP28238.1 KL205624 KFV72230.1 OUUW01000010 SPP86193.1 GEDC01022833 JAS14465.1 JU174157 GBEX01000681 AFJ49683.1 JAI13879.1 GAAZ01000499 GBKC01000707 GBKD01000453 JAA97444.1 JAG45363.1 JAG47165.1 CAQQ02060992 GEBQ01029129 GEBQ01019242 JAT10848.1 JAT20735.1 GBSH01000642 JAG68383.1 KQ419989 KOF81806.1 KK501764 KFP17527.1 KL218474 KFP05688.1 GBSI01000528 JAC95968.1 KL354027 KFZ60847.1 KL384202 KFP89337.1 IACM01108630 IACM01108631 LAB33798.1 AMQM01002998 KB095959 ESO09582.1 GDAY02000469 JAV50944.1 KK381991 KFV48978.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
UP000000673
+ More
UP000075920 UP000075885 UP000069272 UP000005204 UP000075902 UP000075886 UP000075881 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075901 UP000076408 UP000075900 UP000007062 UP000075884 UP000075880 UP000069940 UP000249989 UP000008820 UP000030765 UP000030742 UP000092460 UP000002320 UP000019118 UP000007266 UP000092461 UP000092444 UP000078200 UP000092445 UP000095301 UP000009046 UP000027135 UP000007110 UP000235965 UP000007648 UP000092443 UP000192223 UP000002280 UP000183832 UP000001645 UP000053858 UP000054081 UP000053286 UP000053283 UP000000539 UP000001070 UP000007801 UP000007267 UP000001811 UP000053584 UP000261360 UP000261420 UP000268350 UP000015102 UP000053454 UP000053119 UP000007303 UP000054308 UP000192221 UP000015101 UP000192220
UP000075920 UP000075885 UP000069272 UP000005204 UP000075902 UP000075886 UP000075881 UP000076407 UP000075903 UP000075882 UP000075840 UP000075883 UP000075901 UP000076408 UP000075900 UP000007062 UP000075884 UP000075880 UP000069940 UP000249989 UP000008820 UP000030765 UP000030742 UP000092460 UP000002320 UP000019118 UP000007266 UP000092461 UP000092444 UP000078200 UP000092445 UP000095301 UP000009046 UP000027135 UP000007110 UP000235965 UP000007648 UP000092443 UP000192223 UP000002280 UP000183832 UP000001645 UP000053858 UP000054081 UP000053286 UP000053283 UP000000539 UP000001070 UP000007801 UP000007267 UP000001811 UP000053584 UP000261360 UP000261420 UP000268350 UP000015102 UP000053454 UP000053119 UP000007303 UP000054308 UP000192221 UP000015101 UP000192220
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
Q2F618
A0A2A4JTR9
A0A212ELW7
S4P7E0
A0A2H1VIK9
I4DKP8
+ More
A0A194RHV6 A0A194QAH8 A0A0L7KZW2 A0A2M3ZJZ4 A0A2M4C0I5 W5JGC2 A0A2M4AXW4 A0A182VUG3 B5M0Q9 A0A182PE83 A0A182F3J8 A0A2M4C0F2 H9IVD2 A0A182U6I1 A0A182QY57 A0A182K4V4 A0A182WW33 A0A182VMX0 A0A182L6D7 A0A182HGH8 A0A182MQ98 A0A182T686 A0A182YDR1 A0A182RIP8 A0A1S4H0J1 Q7PVJ3 A0A182NU43 A0A182IY95 A0A182H2A0 Q179F4 A0A0K8TKX8 A0A084WFZ7 U4UUI5 A0A1B0C1H0 A0A1Q3FEH8 B0WFF6 N6UKA8 D2A0V1 A0A1B0CN05 A0A1B0GA75 A0A1A9UPP5 A0A1A9ZP19 T1PK64 A0A1L8DM28 A0A1Y1JWA6 E0VM15 A0A067QNL7 W4ZKD9 A0A2J7PFI5 A0A087V299 G3WRI8 A0A1A9Y571 A0A2J7PFI0 A0A1W4W857 F6WWN8 A0A1J1HTY8 G1N1G4 A0A0A0A700 A0A093NCW4 A0A087QQR1 A0A093T696 A0A091X492 A0A1W4WIG3 F1NM71 B4JBM0 B3N008 K7FX63 G1SWQ3 A0A091JWD2 A0A093GYM0 A0A3B4YHY7 A0A3B4TWQ3 A0A3B0JYP5 A0A1B6CM56 J3S8D8 T1DCW4 T1GD07 A0A1B6LAR5 A0A0B8RZB0 A0A0L8GXM8 A0A091JAB0 H3CU91 A0A091IDW2 A0A098LZR9 A0A1W4W359 A0A094KU40 A0A091NJT5 A0A2D4ML54 T1G1D6 A0A2I4B175 A0A1W7RIH0 A0A3B4X2C1 A0A3B4TX62 A0A093H2L8
A0A194RHV6 A0A194QAH8 A0A0L7KZW2 A0A2M3ZJZ4 A0A2M4C0I5 W5JGC2 A0A2M4AXW4 A0A182VUG3 B5M0Q9 A0A182PE83 A0A182F3J8 A0A2M4C0F2 H9IVD2 A0A182U6I1 A0A182QY57 A0A182K4V4 A0A182WW33 A0A182VMX0 A0A182L6D7 A0A182HGH8 A0A182MQ98 A0A182T686 A0A182YDR1 A0A182RIP8 A0A1S4H0J1 Q7PVJ3 A0A182NU43 A0A182IY95 A0A182H2A0 Q179F4 A0A0K8TKX8 A0A084WFZ7 U4UUI5 A0A1B0C1H0 A0A1Q3FEH8 B0WFF6 N6UKA8 D2A0V1 A0A1B0CN05 A0A1B0GA75 A0A1A9UPP5 A0A1A9ZP19 T1PK64 A0A1L8DM28 A0A1Y1JWA6 E0VM15 A0A067QNL7 W4ZKD9 A0A2J7PFI5 A0A087V299 G3WRI8 A0A1A9Y571 A0A2J7PFI0 A0A1W4W857 F6WWN8 A0A1J1HTY8 G1N1G4 A0A0A0A700 A0A093NCW4 A0A087QQR1 A0A093T696 A0A091X492 A0A1W4WIG3 F1NM71 B4JBM0 B3N008 K7FX63 G1SWQ3 A0A091JWD2 A0A093GYM0 A0A3B4YHY7 A0A3B4TWQ3 A0A3B0JYP5 A0A1B6CM56 J3S8D8 T1DCW4 T1GD07 A0A1B6LAR5 A0A0B8RZB0 A0A0L8GXM8 A0A091JAB0 H3CU91 A0A091IDW2 A0A098LZR9 A0A1W4W359 A0A094KU40 A0A091NJT5 A0A2D4ML54 T1G1D6 A0A2I4B175 A0A1W7RIH0 A0A3B4X2C1 A0A3B4TX62 A0A093H2L8
PDB
5D43
E-value=2.17776e-43,
Score=437
Ontologies
GO
PANTHER
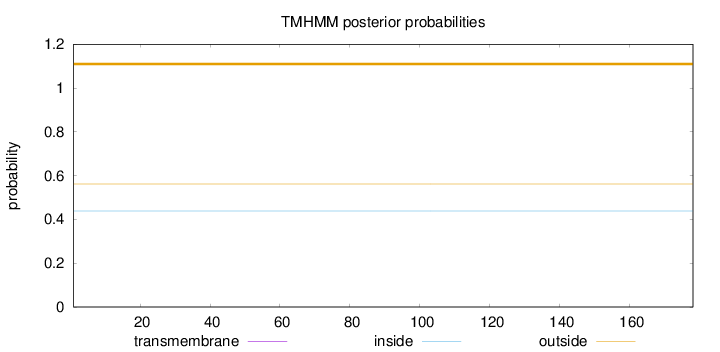
Topology
Length:
178
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.43880
outside
1 - 178
Population Genetic Test Statistics
Pi
240.099508
Theta
199.331758
Tajima's D
0.488769
CLR
0
CSRT
0.506924653767312
Interpretation
Uncertain
