Gene
KWMTBOMO07836
Pre Gene Modal
BGIBMGA000934
Annotation
PREDICTED:_carbonic_anhydrase-related_protein_10_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 1.693 Nuclear Reliability : 1.269
Sequence
CDS
ATGAAGCTAGGAGATTTATCAAATCCGGAACTAAGGGTGCTGACTGAAGAACTTGAGAACATAAAGTACGGGGGTGCTGAAATGCCGGTAAACAAACTATCCGTACGTGGTCTGCTTCCCGATACGGACTACTACATGACGTATGATGGTTCAACCACGGCGCCTGCGTGCTTTGAAACCGTCACCTGGATTATTGTCAACAAACCAATTTACATCACTAAACAACAGCTCCACGGGCTGCGGAGGTTGATGCAGGGCGACGCGAAGCACCCGAAGGCACCCCTCGGCAACAACTTCAGACCACCCCAGCCCCTGCACCACAGGGCTGTTCGAACTAACATTGACTTTGACTTAAACAAGTATCCTGGAAAAACTTGCCCCAGCATGCAAAGAAACATGCATTACAAAGCTAAAAGCTGGACCCAGTACTGA
Protein
MKLGDLSNPELRVLTEELENIKYGGAEMPVNKLSVRGLLPDTDYYMTYDGSTTAPACFETVTWIIVNKPIYITKQQLHGLRRLMQGDAKHPKAPLGNNFRPPQPLHHRAVRTNIDFDLNKYPGKTCPSMQRNMHYKAKSWTQY
Summary
Similarity
Belongs to the alpha-carbonic anhydrase family.
Uniprot
A0A0L7KU05
A0A2A4JPU0
A0A2H1WYE8
A0A194Q9A6
A0A194RLP6
A0A212EYD6
+ More
H9IUK4 A0A1Y1N5R7 A0A1B6KE88 D2A0V8 E0VXZ0 A0A1B6DUA5 B4GWS9 B5DNR6 A0A3B0JZ62 W8BXM4 A0A0A9VU51 B4NE41 A0A1J1HGW3 B3MZF5 A0A1W4VG60 A0A1S3D1H3 B4JIU2 A0A2M4CHF5 B4R765 A0A1I8PB55 B4M267 B4PWZ2 Q9W316 A0A182Q791 B3NT45 B4L2R4 A0A0M4F8I8 A0A1I8MPX8 A0A1A9UED1 A0A182PVM5 A0A182MCF4 A0A182X7I2 A0A182TDG2 A0A1B0G4R7 A0A182FK99 A0A182I9V8 A0A182T9V4 Q17NQ5 B4IDJ4 A0A1S4G9S7 Q7QEE7 A0A182XY80 A0A336M8T2 A0A182W688 A0A087ZUM2 A0A084VVD4 A0A2S2PHG9 A0A158P0L6 A0A0L0C303 A0A0L7R349 K7ISF5 A0A2S2NLE3 A0A026WQ71 A0A2H8TRT8 J9KYZ2 E9ISZ9 A0A226E5S8 A0A154P7Z8 A0A195CRE5 A0A232EJ82 A0A182VAN7 A0A182MZZ3 A0A195F2A3 W5JD04 T1JF35 A0A182JMP5 A0A182I9R9 A0A195BFI7 A0A2J7RTI5 A0A182L6T3 A0A182U066 A0A224XUC8 A0A3L8DXW1 A0A310SG29 A0A2M4CK18 A0A182PTG1 A0A0K8ST31 A0A182J6K1 A0A0Q9WW22 A0A1I8P8P1 A0A0M4FB19 A0A182QJE7 A0A1W4VK37 A0A1A9XXS2 A0A3B0JX60 B5DMT7 B4GVV9 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 B0XKW2 A0A0L0BRC4 W8C9C5
H9IUK4 A0A1Y1N5R7 A0A1B6KE88 D2A0V8 E0VXZ0 A0A1B6DUA5 B4GWS9 B5DNR6 A0A3B0JZ62 W8BXM4 A0A0A9VU51 B4NE41 A0A1J1HGW3 B3MZF5 A0A1W4VG60 A0A1S3D1H3 B4JIU2 A0A2M4CHF5 B4R765 A0A1I8PB55 B4M267 B4PWZ2 Q9W316 A0A182Q791 B3NT45 B4L2R4 A0A0M4F8I8 A0A1I8MPX8 A0A1A9UED1 A0A182PVM5 A0A182MCF4 A0A182X7I2 A0A182TDG2 A0A1B0G4R7 A0A182FK99 A0A182I9V8 A0A182T9V4 Q17NQ5 B4IDJ4 A0A1S4G9S7 Q7QEE7 A0A182XY80 A0A336M8T2 A0A182W688 A0A087ZUM2 A0A084VVD4 A0A2S2PHG9 A0A158P0L6 A0A0L0C303 A0A0L7R349 K7ISF5 A0A2S2NLE3 A0A026WQ71 A0A2H8TRT8 J9KYZ2 E9ISZ9 A0A226E5S8 A0A154P7Z8 A0A195CRE5 A0A232EJ82 A0A182VAN7 A0A182MZZ3 A0A195F2A3 W5JD04 T1JF35 A0A182JMP5 A0A182I9R9 A0A195BFI7 A0A2J7RTI5 A0A182L6T3 A0A182U066 A0A224XUC8 A0A3L8DXW1 A0A310SG29 A0A2M4CK18 A0A182PTG1 A0A0K8ST31 A0A182J6K1 A0A0Q9WW22 A0A1I8P8P1 A0A0M4FB19 A0A182QJE7 A0A1W4VK37 A0A1A9XXS2 A0A3B0JX60 B5DMT7 B4GVV9 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 B0XKW2 A0A0L0BRC4 W8C9C5
Pubmed
26227816
26354079
22118469
19121390
28004739
18362917
+ More
19820115 20566863 17994087 15632085 24495485 25401762 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25315136 17510324 12364791 25244985 24438588 21347285 26108605 20075255 24508170 21282665 28648823 20920257 23761445 20966253 30249741
19820115 20566863 17994087 15632085 24495485 25401762 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25315136 17510324 12364791 25244985 24438588 21347285 26108605 20075255 24508170 21282665 28648823 20920257 23761445 20966253 30249741
EMBL
JTDY01005692
KOB66743.1
NWSH01000817
PCG74067.1
ODYU01011976
SOQ58027.1
+ More
KQ459299 KPJ02004.1 KQ460205 KPJ16906.1 AGBW02011532 OWR46506.1 BABH01000916 BABH01000917 GEZM01014016 JAV92220.1 GEBQ01030214 JAT09763.1 KQ971338 EFA01627.2 DS235842 EEB18246.1 GEDC01008055 JAS29243.1 CH479194 EDW27163.1 CH379065 EDY73168.1 OUUW01000003 SPP78011.1 GAMC01000420 JAC06136.1 GBHO01044370 JAF99233.1 CH964239 EDW82010.2 CVRI01000004 CRK87256.1 CH902635 EDV33756.2 CH916370 EDW00539.1 GGFL01000598 MBW64776.1 CM000366 EDX17534.1 CH940651 EDW65771.1 CM000162 EDX02876.1 AE014298 AY051620 AAF46521.2 AAK93044.1 AFH07307.1 AXCN02001338 AXCN02001339 CH954180 EDV46225.1 CH933810 EDW07862.1 KRF94246.1 KRF94247.1 KRF94248.1 CP012528 ALC48332.1 AXCM01015173 CCAG010009368 CCAG010009369 APCN01001053 CH477197 EAT48348.1 CH480830 EDW45652.1 AAAB01008847 EAA06857.3 UFQS01000677 UFQT01000677 SSX06104.1 SSX26460.1 ATLV01017171 ATLV01017172 ATLV01017173 KE525157 KFB41928.1 GGMR01016260 MBY28879.1 ADTU01005290 ADTU01005291 JRES01001070 KNC25794.1 KQ414663 KOC65310.1 GGMR01005372 MBY17991.1 KK107139 EZA57846.1 GFXV01005100 MBW16905.1 ABLF02029015 GL765434 EFZ16295.1 LNIX01000006 OXA52424.1 KQ434839 KZC08056.1 KQ977394 KYN03067.1 NNAY01004105 OXU18361.1 KQ981856 KYN34511.1 ADMH02001740 ETN61238.1 JH432139 KQ976490 KYM83345.1 NEVH01000002 PNF44141.1 GFTR01004875 JAW11551.1 QOIP01000003 RLU24779.1 KQ768090 OAD53215.1 GGFL01001524 MBW65702.1 GBRD01009473 JAG56351.1 KRF99656.1 ALC49720.1 AXCN02001216 SPP78339.1 CH379064 EDY73119.1 CH479193 EDW26804.1 EDX02345.2 CH480865 EDW53653.1 EDV47494.1 BT024405 AAF46274.4 ABC86467.1 AHN59457.1 DS234059 EDS32747.1 JRES01001480 KNC22587.1 GAMC01002279 JAC04277.1
KQ459299 KPJ02004.1 KQ460205 KPJ16906.1 AGBW02011532 OWR46506.1 BABH01000916 BABH01000917 GEZM01014016 JAV92220.1 GEBQ01030214 JAT09763.1 KQ971338 EFA01627.2 DS235842 EEB18246.1 GEDC01008055 JAS29243.1 CH479194 EDW27163.1 CH379065 EDY73168.1 OUUW01000003 SPP78011.1 GAMC01000420 JAC06136.1 GBHO01044370 JAF99233.1 CH964239 EDW82010.2 CVRI01000004 CRK87256.1 CH902635 EDV33756.2 CH916370 EDW00539.1 GGFL01000598 MBW64776.1 CM000366 EDX17534.1 CH940651 EDW65771.1 CM000162 EDX02876.1 AE014298 AY051620 AAF46521.2 AAK93044.1 AFH07307.1 AXCN02001338 AXCN02001339 CH954180 EDV46225.1 CH933810 EDW07862.1 KRF94246.1 KRF94247.1 KRF94248.1 CP012528 ALC48332.1 AXCM01015173 CCAG010009368 CCAG010009369 APCN01001053 CH477197 EAT48348.1 CH480830 EDW45652.1 AAAB01008847 EAA06857.3 UFQS01000677 UFQT01000677 SSX06104.1 SSX26460.1 ATLV01017171 ATLV01017172 ATLV01017173 KE525157 KFB41928.1 GGMR01016260 MBY28879.1 ADTU01005290 ADTU01005291 JRES01001070 KNC25794.1 KQ414663 KOC65310.1 GGMR01005372 MBY17991.1 KK107139 EZA57846.1 GFXV01005100 MBW16905.1 ABLF02029015 GL765434 EFZ16295.1 LNIX01000006 OXA52424.1 KQ434839 KZC08056.1 KQ977394 KYN03067.1 NNAY01004105 OXU18361.1 KQ981856 KYN34511.1 ADMH02001740 ETN61238.1 JH432139 KQ976490 KYM83345.1 NEVH01000002 PNF44141.1 GFTR01004875 JAW11551.1 QOIP01000003 RLU24779.1 KQ768090 OAD53215.1 GGFL01001524 MBW65702.1 GBRD01009473 JAG56351.1 KRF99656.1 ALC49720.1 AXCN02001216 SPP78339.1 CH379064 EDY73119.1 CH479193 EDW26804.1 EDX02345.2 CH480865 EDW53653.1 EDV47494.1 BT024405 AAF46274.4 ABC86467.1 AHN59457.1 DS234059 EDS32747.1 JRES01001480 KNC22587.1 GAMC01002279 JAC04277.1
Proteomes
UP000037510
UP000218220
UP000053268
UP000053240
UP000007151
UP000005204
+ More
UP000007266 UP000009046 UP000008744 UP000001819 UP000268350 UP000007798 UP000183832 UP000007801 UP000192221 UP000079169 UP000001070 UP000000304 UP000095300 UP000008792 UP000002282 UP000000803 UP000075886 UP000008711 UP000009192 UP000092553 UP000095301 UP000078200 UP000075885 UP000075883 UP000076407 UP000075902 UP000092444 UP000069272 UP000075840 UP000075901 UP000008820 UP000001292 UP000007062 UP000076408 UP000075920 UP000005203 UP000030765 UP000005205 UP000037069 UP000053825 UP000002358 UP000053097 UP000007819 UP000198287 UP000076502 UP000078542 UP000215335 UP000075903 UP000075884 UP000078541 UP000000673 UP000075880 UP000078540 UP000235965 UP000075882 UP000279307 UP000092443 UP000002320
UP000007266 UP000009046 UP000008744 UP000001819 UP000268350 UP000007798 UP000183832 UP000007801 UP000192221 UP000079169 UP000001070 UP000000304 UP000095300 UP000008792 UP000002282 UP000000803 UP000075886 UP000008711 UP000009192 UP000092553 UP000095301 UP000078200 UP000075885 UP000075883 UP000076407 UP000075902 UP000092444 UP000069272 UP000075840 UP000075901 UP000008820 UP000001292 UP000007062 UP000076408 UP000075920 UP000005203 UP000030765 UP000005205 UP000037069 UP000053825 UP000002358 UP000053097 UP000007819 UP000198287 UP000076502 UP000078542 UP000215335 UP000075903 UP000075884 UP000078541 UP000000673 UP000075880 UP000078540 UP000235965 UP000075882 UP000279307 UP000092443 UP000002320
Interpro
Gene 3D
ProteinModelPortal
A0A0L7KU05
A0A2A4JPU0
A0A2H1WYE8
A0A194Q9A6
A0A194RLP6
A0A212EYD6
+ More
H9IUK4 A0A1Y1N5R7 A0A1B6KE88 D2A0V8 E0VXZ0 A0A1B6DUA5 B4GWS9 B5DNR6 A0A3B0JZ62 W8BXM4 A0A0A9VU51 B4NE41 A0A1J1HGW3 B3MZF5 A0A1W4VG60 A0A1S3D1H3 B4JIU2 A0A2M4CHF5 B4R765 A0A1I8PB55 B4M267 B4PWZ2 Q9W316 A0A182Q791 B3NT45 B4L2R4 A0A0M4F8I8 A0A1I8MPX8 A0A1A9UED1 A0A182PVM5 A0A182MCF4 A0A182X7I2 A0A182TDG2 A0A1B0G4R7 A0A182FK99 A0A182I9V8 A0A182T9V4 Q17NQ5 B4IDJ4 A0A1S4G9S7 Q7QEE7 A0A182XY80 A0A336M8T2 A0A182W688 A0A087ZUM2 A0A084VVD4 A0A2S2PHG9 A0A158P0L6 A0A0L0C303 A0A0L7R349 K7ISF5 A0A2S2NLE3 A0A026WQ71 A0A2H8TRT8 J9KYZ2 E9ISZ9 A0A226E5S8 A0A154P7Z8 A0A195CRE5 A0A232EJ82 A0A182VAN7 A0A182MZZ3 A0A195F2A3 W5JD04 T1JF35 A0A182JMP5 A0A182I9R9 A0A195BFI7 A0A2J7RTI5 A0A182L6T3 A0A182U066 A0A224XUC8 A0A3L8DXW1 A0A310SG29 A0A2M4CK18 A0A182PTG1 A0A0K8ST31 A0A182J6K1 A0A0Q9WW22 A0A1I8P8P1 A0A0M4FB19 A0A182QJE7 A0A1W4VK37 A0A1A9XXS2 A0A3B0JX60 B5DMT7 B4GVV9 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 B0XKW2 A0A0L0BRC4 W8C9C5
H9IUK4 A0A1Y1N5R7 A0A1B6KE88 D2A0V8 E0VXZ0 A0A1B6DUA5 B4GWS9 B5DNR6 A0A3B0JZ62 W8BXM4 A0A0A9VU51 B4NE41 A0A1J1HGW3 B3MZF5 A0A1W4VG60 A0A1S3D1H3 B4JIU2 A0A2M4CHF5 B4R765 A0A1I8PB55 B4M267 B4PWZ2 Q9W316 A0A182Q791 B3NT45 B4L2R4 A0A0M4F8I8 A0A1I8MPX8 A0A1A9UED1 A0A182PVM5 A0A182MCF4 A0A182X7I2 A0A182TDG2 A0A1B0G4R7 A0A182FK99 A0A182I9V8 A0A182T9V4 Q17NQ5 B4IDJ4 A0A1S4G9S7 Q7QEE7 A0A182XY80 A0A336M8T2 A0A182W688 A0A087ZUM2 A0A084VVD4 A0A2S2PHG9 A0A158P0L6 A0A0L0C303 A0A0L7R349 K7ISF5 A0A2S2NLE3 A0A026WQ71 A0A2H8TRT8 J9KYZ2 E9ISZ9 A0A226E5S8 A0A154P7Z8 A0A195CRE5 A0A232EJ82 A0A182VAN7 A0A182MZZ3 A0A195F2A3 W5JD04 T1JF35 A0A182JMP5 A0A182I9R9 A0A195BFI7 A0A2J7RTI5 A0A182L6T3 A0A182U066 A0A224XUC8 A0A3L8DXW1 A0A310SG29 A0A2M4CK18 A0A182PTG1 A0A0K8ST31 A0A182J6K1 A0A0Q9WW22 A0A1I8P8P1 A0A0M4FB19 A0A182QJE7 A0A1W4VK37 A0A1A9XXS2 A0A3B0JX60 B5DMT7 B4GVV9 B4Q0S4 B4IL36 B3NTT1 Q9W3P7 B0XKW2 A0A0L0BRC4 W8C9C5
PDB
1Z97
E-value=3.74259e-17,
Score=209
Ontologies
PANTHER
Topology
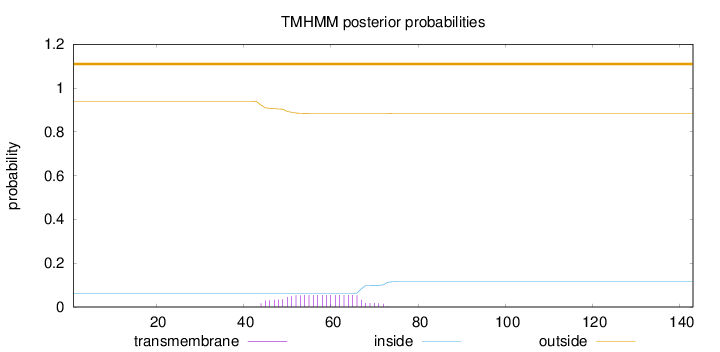
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21091
Exp number, first 60 AAs:
0.7627
Total prob of N-in:
0.06168
outside
1 - 143
Population Genetic Test Statistics
Pi
168.147741
Theta
152.342994
Tajima's D
0.5915
CLR
0
CSRT
0.546122693865307
Interpretation
Possibly Positive selection
