Gene
KWMTBOMO07830
Pre Gene Modal
BGIBMGA000932
Annotation
PREDICTED:_serine/threonine-protein_phosphatase_rdgC_[Bombyx_mori]
Full name
Serine/threonine-protein phosphatase
+ More
Serine/threonine-protein phosphatase with EF-hands
Serine/threonine-protein phosphatase rdgC
Serine/threonine-protein phosphatase with EF-hands
Serine/threonine-protein phosphatase rdgC
Alternative Name
Retinal degeneration C protein
Location in the cell
Nuclear Reliability : 2.096
Sequence
CDS
ATGTTAGCCGCAGCCATTCCTAAATCCCGCGAAAAAGTGCCAAGATGCAATCGAAAGCGAAGAGATAAAAGCCACAGAGTAAGGGCCAAGAGTCCCACGATGTTTTCTTTGGCTGAGGGTTGTGGGTGCGTTAGTTTTCGTGGCGGTCGACGTACTCGCTCCCAAGGAGATGATTCTTCATCATGTCGGAGCTCAGAAATAGAACAAGAGGTTCCCCAAAGATCGGGTAAAAGACTACTCAATGGCGAATATACCATGAAAGCTTTGGTACCCTTCTTGAAATTTTGGAGAAGAGACGATGTTAGTATGACTAAAGCTGAAAGAACTATGAAAGCCGCCATTTTAATTCAGCAGTGGTACAGGCGTTATTTAGCGCGAATGGAAGTGAGAAGGCGTTATACCTGGACCATATTTCAGAACATTGAATATGCGGGCGAACATGATCAAGTGAAGCTGTATAATTTTTTCAACGCACTGCTCAGCCATATGATGGATACGAATGGGCGGCCCTCCTCCGAGCGTACCTCGAGAACAGCTTCCGTCTTGGAACAAGTTTTTTCCGAAGAGTCTGACGAAGGAGGGGGCGTCGATGAGACCGCTCCCCCGAAGCCTTCACGTGGCCCCGACGTTGAATTCCCGCTACAAAGAAAGGATATCGACCTATTGATTGAGGCATTCAGAAAGAAAAAGCATCGTTTACATCCAAAGCACGTAGCGCAGATACTGAAAGAGGCCACACAAGTATTGAAAAGGTTACCCAATATCAATGTGGCTACCACATCTATATCTAGCCAGATCACTGTTTGCGGAGATCTCCATGGAAAACTAGACGATCTACTCGTCATATTTCACAAGAATGGGCTGCCATCTCCAGAAAATCCGTATATATTTAACGGAGACTTTGTGGACAGAGGCAAGAAGGGTCTAGAAGTGTTTCTTCTGCTGTTAGCGTGCATGCTCGCTTTTCCCGGTGGTGTTTTCTTGAATCGCGGGAATCACGAAGATCTCATTATGAATTCGAGGTACGGCTTCATCAAAGAAGTTCACATGAAGTACAGGCACAATGCTGAGCGTCTTTTGAAACTGATTGAGTGCGTGTATCGGTGGCTTCCACTCGGGACCATCATAAATAATCGAGTTTTCGTTGTCCACGGAGGCATCTCGGATACTACAGACCTAGATATGATTCGGAGCCTTGAAAGGGATAAGTACGTGTCGCTTCTGCGGCCGCCGGTGATCGAGGGCTCAACGGCCGCGGAGTCGATAGATAAGGTGGAGTGGAAGCAGGTGTTTGATATACTGTGGTCGGATCCACAATGCATCGGAGGATGTGTGGCAAATGCGCTTCGAGGAGCAGGAACCTACTTCGGCCCTGATGTAACAGCGTCGTTCCTCCAGAAGAATAAGCTGAGCTTCATAATTAGAAGTCACGAGTGCAAGCCCGGTGGATACGAAATCATGCATGGCGGTAATGTAATTACAATTTTCTCGGCGTCGAACTATTACGAGTTGGGTTCCAACAAGGGCGCTTACCTGAAACTATGCGGGAACTCGTTGGAGCGCCAGTTCGTGCAATACACGGCGGCCGTGTCTCGCACGCGGCGTCTCACCTTCAGGCAGCGGGTTGGCCTTGTCGAATCGTCCGCCATGAGGGAGCTACACAATCAGATTATGATGGCCAGGAGATCTCTGGAAGCTACTTTCCGTAGCTTGGACTCGGAGCAAAGTGGTTTCATATCTATAAGCGATTGGTGTCAAGCCATGGAGGAGACAACACACCTCGGTCTCCCGTGGAGGATGCTTAAAGACAAGCTTGTTCGCACAGATCCCGAGACACGCAAAGTACGTTACATGGATACATTCGAACTCAACACCAATAGGAACGGACGAAAATCTGACACGCTGACAGTCGTCGAGACTTTGTATAAAAATCAAAGAAGCTTGGAGGCGATCTTCAAGATATTGGATAAAGATAATTCTGGTTACATAACGTTGGAAGAATTTTCCGAGGCTTGCCAGCTGATCGGCAAGTACATGCCGAATCCGATGACGCAGGAGCAGCTCGTCGACATCTGTCGGCTCATGGACATCAACAAGGACGGCCTGGTTGATCTCAACGAGTTCCTCGAGAGTTTCCGTCTCGCCGAAAAGAGTTTACACGAGCAGGAAAGGATGTACGAAGACATGCAGGCTACCTGA
Protein
MLAAAIPKSREKVPRCNRKRRDKSHRVRAKSPTMFSLAEGCGCVSFRGGRRTRSQGDDSSSCRSSEIEQEVPQRSGKRLLNGEYTMKALVPFLKFWRRDDVSMTKAERTMKAAILIQQWYRRYLARMEVRRRYTWTIFQNIEYAGEHDQVKLYNFFNALLSHMMDTNGRPSSERTSRTASVLEQVFSEESDEGGGVDETAPPKPSRGPDVEFPLQRKDIDLLIEAFRKKKHRLHPKHVAQILKEATQVLKRLPNINVATTSISSQITVCGDLHGKLDDLLVIFHKNGLPSPENPYIFNGDFVDRGKKGLEVFLLLLACMLAFPGGVFLNRGNHEDLIMNSRYGFIKEVHMKYRHNAERLLKLIECVYRWLPLGTIINNRVFVVHGGISDTTDLDMIRSLERDKYVSLLRPPVIEGSTAAESIDKVEWKQVFDILWSDPQCIGGCVANALRGAGTYFGPDVTASFLQKNKLSFIIRSHECKPGGYEIMHGGNVITIFSASNYYELGSNKGAYLKLCGNSLERQFVQYTAAVSRTRRLTFRQRVGLVESSAMRELHNQIMMARRSLEATFRSLDSEQSGFISISDWCQAMEETTHLGLPWRMLKDKLVRTDPETRKVRYMDTFELNTNRNGRKSDTLTVVETLYKNQRSLEAIFKILDKDNSGYITLEEFSEACQLIGKYMPNPMTQEQLVDICRLMDINKDGLVDLNEFLESFRLAEKSLHEQERMYEDMQAT
Summary
Description
Phosphatase required to prevent light-induced retinal degeneration.
Catalytic Activity
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
Cofactor
Mn(2+)
Similarity
Belongs to the PPP phosphatase family.
Keywords
Calcium
Complete proteome
Hydrolase
Magnesium
Manganese
Metal-binding
Protein phosphatase
Reference proteome
Repeat
Sensory transduction
Vision
Feature
chain Serine/threonine-protein phosphatase rdgC
Uniprot
A0A194RLP1
H9IUK2
A0A194PJL2
A0A212EZK5
A0A1W4WIT1
A0A1S4F4H5
+ More
D2A162 W5JP23 Q17FS3 A0A084WI84 A0A182WR55 A0A182HNP8 A0A182WF17 A0A182RFE2 A0A182NPL6 A0A182FTV4 A0A182PU84 Q7Q2Z9 A0A336KR11 A0A182G6D2 A0A182IJK1 A0A1B6DP80 A0A1B6C198 A0A182V902 A0A0A9XJD6 A0A182MN75 A0A0L0BQ75 A0A0T6AY67 A0A1I8QDX9 E0VT76 B0W8E1 A0A182K8J4 A0A1B0G902 A0A336KZ48 U4US14 A0A1A9WRH1 W8ANN1 A0A1A9VSD6 A0A1A9Z5T7 A0A1B0G8C3 A0A1A9Z4V7 A0A1A9UQS4 A0A0Q5U6J1 A0A1B0B3L8 B4PFC5 A0A0J9RZW2 A0A1B0G581 A0A1W4VUR9 A0A0P8XTK4 A0A3B0K9C5 A0A1A9Z2Z5 A0A0Q5UKB6 A0A0R1E263 B3NE58 A0A0J9RYJ5 A0A0R1E3V8 B4IA68 A0A0J9UPV3 B4MKU9 P40421 A0A0P8XTZ4 A0A0R3P3C9 B3M4R3 A0A3B0KHY3 A0A0Q9WVD8 A0A0R3P3E4 A0A0R3P3H1 A0A0R3P7M8 B5DPA9 A0A1J1IGC9 A0A0Q9XDH4 B4IYR3 B4LG02 A0A0Q9XQA2 B4KV71 A0A0M4EZY5 A0A1A9Y029 B4GR08 A0A2T7PP07 A0A224Z6H9 A0A131YUK5 A0A369SL60 A0A044UXV0 A0A090LKG0 A7RNB7 A0A0N5BXR6 A0A0K0EJQ2 A0A2B4SS40 A0A1S3HU17 A0A0K0F6X9 A0A1I7VS43
D2A162 W5JP23 Q17FS3 A0A084WI84 A0A182WR55 A0A182HNP8 A0A182WF17 A0A182RFE2 A0A182NPL6 A0A182FTV4 A0A182PU84 Q7Q2Z9 A0A336KR11 A0A182G6D2 A0A182IJK1 A0A1B6DP80 A0A1B6C198 A0A182V902 A0A0A9XJD6 A0A182MN75 A0A0L0BQ75 A0A0T6AY67 A0A1I8QDX9 E0VT76 B0W8E1 A0A182K8J4 A0A1B0G902 A0A336KZ48 U4US14 A0A1A9WRH1 W8ANN1 A0A1A9VSD6 A0A1A9Z5T7 A0A1B0G8C3 A0A1A9Z4V7 A0A1A9UQS4 A0A0Q5U6J1 A0A1B0B3L8 B4PFC5 A0A0J9RZW2 A0A1B0G581 A0A1W4VUR9 A0A0P8XTK4 A0A3B0K9C5 A0A1A9Z2Z5 A0A0Q5UKB6 A0A0R1E263 B3NE58 A0A0J9RYJ5 A0A0R1E3V8 B4IA68 A0A0J9UPV3 B4MKU9 P40421 A0A0P8XTZ4 A0A0R3P3C9 B3M4R3 A0A3B0KHY3 A0A0Q9WVD8 A0A0R3P3E4 A0A0R3P3H1 A0A0R3P7M8 B5DPA9 A0A1J1IGC9 A0A0Q9XDH4 B4IYR3 B4LG02 A0A0Q9XQA2 B4KV71 A0A0M4EZY5 A0A1A9Y029 B4GR08 A0A2T7PP07 A0A224Z6H9 A0A131YUK5 A0A369SL60 A0A044UXV0 A0A090LKG0 A7RNB7 A0A0N5BXR6 A0A0K0EJQ2 A0A2B4SS40 A0A1S3HU17 A0A0K0F6X9 A0A1I7VS43
EC Number
3.1.3.16
Pubmed
EMBL
KQ460205
KPJ16901.1
BABH01000935
KQ459606
KPI91275.1
AGBW02011268
+ More
OWR46920.1 KQ971338 EFA01582.2 ADMH02000929 ETN64645.1 CH477268 EAT45428.1 ATLV01023923 KE525347 KFB49928.1 AAAB01008966 EAA12933.4 UFQS01000791 UFQT01000791 SSX06909.1 SSX27253.1 JXUM01007767 JXUM01007768 JXUM01007769 JXUM01007770 JXUM01007771 JXUM01007772 KQ560247 KXJ83583.1 GEDC01009827 JAS27471.1 GEDC01030006 JAS07292.1 GBHO01023525 GBHO01023524 GDHC01015922 JAG20079.1 JAG20080.1 JAQ02707.1 AXCM01005116 JRES01001542 KNC22133.1 LJIG01022522 KRT80131.1 DS235760 EEB16582.1 DS231858 EDS38870.1 CCAG010002954 CCAG010002955 CCAG010002956 UFQS01001279 UFQT01001279 SSX10107.1 SSX29829.1 KB632337 ERL92855.1 GAMC01016156 GAMC01016155 JAB90400.1 CCAG010000237 CH954178 KQS44297.1 JXJN01007898 JXJN01007899 CM000159 EDW95211.1 CM002912 KMZ00725.1 CCAG010004851 CH902618 KPU78056.1 OUUW01000009 SPP84720.1 KQS44298.1 KRK02291.1 EDV52482.1 KMZ00727.1 KRK02292.1 CH480826 EDW44181.1 KMZ00726.1 CH963847 EDW72874.2 M89628 AE014296 BT015959 KPU78057.1 CH379069 KRT07671.1 EDV39462.1 SPP84721.1 CH940647 KRF84894.1 KRT07672.1 KRT07673.1 KRT07669.1 EDY73443.1 CVRI01000047 CRK98102.1 CH933809 KRG06630.1 CH916366 EDV96600.1 EDW70401.1 KRG06631.1 EDW19411.1 CP012525 ALC44362.1 CH479188 EDW40193.1 PZQS01000003 PVD35141.1 GFPF01011705 MAA22851.1 GEDV01005900 JAP82657.1 NOWV01000006 RDD46763.1 CMVM020000240 LN609529 CEF68045.1 DS469522 EDO47034.1 LSMT01000037 PFX31297.1 JH712149 EJD75329.1
OWR46920.1 KQ971338 EFA01582.2 ADMH02000929 ETN64645.1 CH477268 EAT45428.1 ATLV01023923 KE525347 KFB49928.1 AAAB01008966 EAA12933.4 UFQS01000791 UFQT01000791 SSX06909.1 SSX27253.1 JXUM01007767 JXUM01007768 JXUM01007769 JXUM01007770 JXUM01007771 JXUM01007772 KQ560247 KXJ83583.1 GEDC01009827 JAS27471.1 GEDC01030006 JAS07292.1 GBHO01023525 GBHO01023524 GDHC01015922 JAG20079.1 JAG20080.1 JAQ02707.1 AXCM01005116 JRES01001542 KNC22133.1 LJIG01022522 KRT80131.1 DS235760 EEB16582.1 DS231858 EDS38870.1 CCAG010002954 CCAG010002955 CCAG010002956 UFQS01001279 UFQT01001279 SSX10107.1 SSX29829.1 KB632337 ERL92855.1 GAMC01016156 GAMC01016155 JAB90400.1 CCAG010000237 CH954178 KQS44297.1 JXJN01007898 JXJN01007899 CM000159 EDW95211.1 CM002912 KMZ00725.1 CCAG010004851 CH902618 KPU78056.1 OUUW01000009 SPP84720.1 KQS44298.1 KRK02291.1 EDV52482.1 KMZ00727.1 KRK02292.1 CH480826 EDW44181.1 KMZ00726.1 CH963847 EDW72874.2 M89628 AE014296 BT015959 KPU78057.1 CH379069 KRT07671.1 EDV39462.1 SPP84721.1 CH940647 KRF84894.1 KRT07672.1 KRT07673.1 KRT07669.1 EDY73443.1 CVRI01000047 CRK98102.1 CH933809 KRG06630.1 CH916366 EDV96600.1 EDW70401.1 KRG06631.1 EDW19411.1 CP012525 ALC44362.1 CH479188 EDW40193.1 PZQS01000003 PVD35141.1 GFPF01011705 MAA22851.1 GEDV01005900 JAP82657.1 NOWV01000006 RDD46763.1 CMVM020000240 LN609529 CEF68045.1 DS469522 EDO47034.1 LSMT01000037 PFX31297.1 JH712149 EJD75329.1
Proteomes
UP000053240
UP000005204
UP000053268
UP000007151
UP000192223
UP000007266
+ More
UP000000673 UP000008820 UP000030765 UP000076407 UP000075920 UP000075900 UP000075884 UP000069272 UP000075885 UP000007062 UP000069940 UP000249989 UP000075880 UP000075903 UP000075883 UP000037069 UP000095300 UP000009046 UP000002320 UP000075881 UP000092444 UP000030742 UP000091820 UP000078200 UP000092445 UP000008711 UP000092460 UP000002282 UP000192221 UP000007801 UP000268350 UP000001292 UP000007798 UP000000803 UP000001819 UP000008792 UP000183832 UP000009192 UP000001070 UP000092553 UP000092443 UP000008744 UP000245119 UP000253843 UP000024404 UP000035682 UP000001593 UP000046392 UP000035681 UP000225706 UP000085678 UP000035680 UP000095285
UP000000673 UP000008820 UP000030765 UP000076407 UP000075920 UP000075900 UP000075884 UP000069272 UP000075885 UP000007062 UP000069940 UP000249989 UP000075880 UP000075903 UP000075883 UP000037069 UP000095300 UP000009046 UP000002320 UP000075881 UP000092444 UP000030742 UP000091820 UP000078200 UP000092445 UP000008711 UP000092460 UP000002282 UP000192221 UP000007801 UP000268350 UP000001292 UP000007798 UP000000803 UP000001819 UP000008792 UP000183832 UP000009192 UP000001070 UP000092553 UP000092443 UP000008744 UP000245119 UP000253843 UP000024404 UP000035682 UP000001593 UP000046392 UP000035681 UP000225706 UP000085678 UP000035680 UP000095285
Pfam
Interpro
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
A0A194RLP1
H9IUK2
A0A194PJL2
A0A212EZK5
A0A1W4WIT1
A0A1S4F4H5
+ More
D2A162 W5JP23 Q17FS3 A0A084WI84 A0A182WR55 A0A182HNP8 A0A182WF17 A0A182RFE2 A0A182NPL6 A0A182FTV4 A0A182PU84 Q7Q2Z9 A0A336KR11 A0A182G6D2 A0A182IJK1 A0A1B6DP80 A0A1B6C198 A0A182V902 A0A0A9XJD6 A0A182MN75 A0A0L0BQ75 A0A0T6AY67 A0A1I8QDX9 E0VT76 B0W8E1 A0A182K8J4 A0A1B0G902 A0A336KZ48 U4US14 A0A1A9WRH1 W8ANN1 A0A1A9VSD6 A0A1A9Z5T7 A0A1B0G8C3 A0A1A9Z4V7 A0A1A9UQS4 A0A0Q5U6J1 A0A1B0B3L8 B4PFC5 A0A0J9RZW2 A0A1B0G581 A0A1W4VUR9 A0A0P8XTK4 A0A3B0K9C5 A0A1A9Z2Z5 A0A0Q5UKB6 A0A0R1E263 B3NE58 A0A0J9RYJ5 A0A0R1E3V8 B4IA68 A0A0J9UPV3 B4MKU9 P40421 A0A0P8XTZ4 A0A0R3P3C9 B3M4R3 A0A3B0KHY3 A0A0Q9WVD8 A0A0R3P3E4 A0A0R3P3H1 A0A0R3P7M8 B5DPA9 A0A1J1IGC9 A0A0Q9XDH4 B4IYR3 B4LG02 A0A0Q9XQA2 B4KV71 A0A0M4EZY5 A0A1A9Y029 B4GR08 A0A2T7PP07 A0A224Z6H9 A0A131YUK5 A0A369SL60 A0A044UXV0 A0A090LKG0 A7RNB7 A0A0N5BXR6 A0A0K0EJQ2 A0A2B4SS40 A0A1S3HU17 A0A0K0F6X9 A0A1I7VS43
D2A162 W5JP23 Q17FS3 A0A084WI84 A0A182WR55 A0A182HNP8 A0A182WF17 A0A182RFE2 A0A182NPL6 A0A182FTV4 A0A182PU84 Q7Q2Z9 A0A336KR11 A0A182G6D2 A0A182IJK1 A0A1B6DP80 A0A1B6C198 A0A182V902 A0A0A9XJD6 A0A182MN75 A0A0L0BQ75 A0A0T6AY67 A0A1I8QDX9 E0VT76 B0W8E1 A0A182K8J4 A0A1B0G902 A0A336KZ48 U4US14 A0A1A9WRH1 W8ANN1 A0A1A9VSD6 A0A1A9Z5T7 A0A1B0G8C3 A0A1A9Z4V7 A0A1A9UQS4 A0A0Q5U6J1 A0A1B0B3L8 B4PFC5 A0A0J9RZW2 A0A1B0G581 A0A1W4VUR9 A0A0P8XTK4 A0A3B0K9C5 A0A1A9Z2Z5 A0A0Q5UKB6 A0A0R1E263 B3NE58 A0A0J9RYJ5 A0A0R1E3V8 B4IA68 A0A0J9UPV3 B4MKU9 P40421 A0A0P8XTZ4 A0A0R3P3C9 B3M4R3 A0A3B0KHY3 A0A0Q9WVD8 A0A0R3P3E4 A0A0R3P3H1 A0A0R3P7M8 B5DPA9 A0A1J1IGC9 A0A0Q9XDH4 B4IYR3 B4LG02 A0A0Q9XQA2 B4KV71 A0A0M4EZY5 A0A1A9Y029 B4GR08 A0A2T7PP07 A0A224Z6H9 A0A131YUK5 A0A369SL60 A0A044UXV0 A0A090LKG0 A7RNB7 A0A0N5BXR6 A0A0K0EJQ2 A0A2B4SS40 A0A1S3HU17 A0A0K0F6X9 A0A1I7VS43
PDB
3H69
E-value=2.84228e-63,
Score=616
Ontologies
GO
Topology
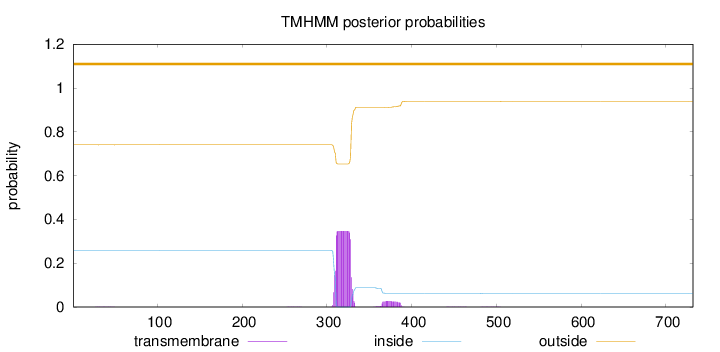
Length:
732
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.50338999999999
Exp number, first 60 AAs:
0.00879
Total prob of N-in:
0.25924
outside
1 - 732
Population Genetic Test Statistics
Pi
252.16079
Theta
186.796423
Tajima's D
0.987717
CLR
0.311684
CSRT
0.656917154142293
Interpretation
Uncertain
