Pre Gene Modal
BGIBMGA000931
Annotation
PREDICTED:_CCR4-NOT_transcription_complex_subunit_7-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.891 Nuclear Reliability : 1.375
Sequence
CDS
ATGAAACAAAAAAGGTGCCCATCAGGAGAAAAGATGCCGGCGACCAGTTTCACCTCTTTACTGTGCAGTGCATTAAAAGAAGAGTGCAGTATTAAAAACGTTTGGAATCATAATCTTCATGAAGAATTTCATGTAATAAGACAGGTTGTCCAGAAATACCATTGGGTGGCCATGGATACTGAATTTCCAGGTGTAGTTGCAAGACCAATTGGTGAATTCAGATCCACTGCAGATTATCAATACCAGCTTTTAAGATGCAATGTGGATCTGTTGCGGATCATACAATTAGGATTAACGTTTATGGACGAAAATGCTAAAACTCCCCCAGGGTGCACAACTTGGCAGTTTAATTTTAAATTCAATCTTCAGGAGGATATGTATGCACAAGACTCGATAGACTTACTACAAAATTCTGGTCTGCAATTCAGGGAACATGAAGAACATGGTATTGAACCCCTTGAATTTGCTGAATTACTAATGTCATCTGGAATTGTACTAATGGATAACATCAACTGGTTGAGTTTTCATTCTGGATATGATTTTGGTTATTTACTTAAGTTGCTTACTGATCAAAACCTACCTCATGAAGAGAATGACTTCTTTGAACGATTGCGTCTATATTTCCCCACCGTGTATGATGTCAAGTACCTGATGAAACTTTGCAAGAATCTTAAAGGTGGCTTACAAGAAGTTGCAGATCAGTTGGAATTGAGGCGAGTAGGTCCCCAACATCAAGCCGGTTCTGACTCACTACTGACTGGCATGGCGTTTTTTAAAATTAAAGAAATATTTTTTGATGGAAACATTGAAAGTACGAGTGGACATCTGTACGGACTTGGAGCTCCATTCACTACTAACGCCAACAACTTCCAGGATAACGTGGATAGTGGAAACTCATCTTGA
Protein
MKQKRCPSGEKMPATSFTSLLCSALKEECSIKNVWNHNLHEEFHVIRQVVQKYHWVAMDTEFPGVVARPIGEFRSTADYQYQLLRCNVDLLRIIQLGLTFMDENAKTPPGCTTWQFNFKFNLQEDMYAQDSIDLLQNSGLQFREHEEHGIEPLEFAELLMSSGIVLMDNINWLSFHSGYDFGYLLKLLTDQNLPHEENDFFERLRLYFPTVYDVKYLMKLCKNLKGGLQEVADQLELRRVGPQHQAGSDSLLTGMAFFKIKEIFFDGNIESTSGHLYGLGAPFTTNANNFQDNVDSGNSS
Summary
Uniprot
A0A1E1WI84
A0A2A4J6V5
A0A2H1WY06
A0A194PEZ7
I4DR32
A0A2A4J6P5
+ More
A0A1E1W6H0 A0A2H1VS46 H9IUK1 A0A212EZJ3 A0A194RHU6 A0A212EZJ4 A0A194Q6F0 I4DKN7 I4DRB7 A0A194QVX4 S4PL71 A0A067RBB7 A0A1B0CUL9 A0A1B6KQM6 A0A1L8DGU4 A0A1L8DGV1 A0A1B6HZF3 A0A1B6GM18 A0A1L8DGR3 A0A336K9F1 A0A1B6BXZ8 A0A336KBA5 A0A0M4ENG0 A0A0Q9XR26 A0A1B6EAQ6 B4L164 A0A0K8TQV2 A0A0Q9WM05 W8ASK5 A0A3B0JPQ5 A0A0P9AKQ5 B4LC30 A0A3B0K245 B3MAG0 W8B455 B4H1W5 A0A034W2B8 A0A0K8WAS3 Q29EE0 A0A0K8TV29 A0A034W6F8 A0A0K8UNG5 A0A0R1E1Z7 A0A0Q5UJ14 B4MLI7 A0A3B0JUS5 B4PG79 B3NH50 Q16VZ2 A0A1W4W603 Q16VZ3 B4IXY9 A0A182GKR0 A0A224XJ77 U4UD40 A0A0P4VJX9 A0A023F5F6 A0A069DSE9 A0A1W4W7P3 J3JV26 A0A084WI87 Q8I0I5 A0A1I8PU18 A0A1I8PU50 V5GRD5 T1P857 Q9VTS4 A0A0L0BZQ3 A0A0A9XQT1 A0A023ES05 A0A1I8N2Q5 A0A1I8PU32 A0A1I8N2Q9 A0A1Y1LSN9 A0A0T6B7Y0 A0A0J9RU39 A0A1I8JT44 Q8IGD6 A0A0A9XKD7 A0A182WR57 A0A182LPR2 Q7Q2Z8 D2A3D8 A0A0J9RTR8 A0A1B0C4U7 A0A0P5W4P6 A0A2P8XSW5 D3TP58 A0A1A9XGF6 A0A1A9ZKN2 A0A1B0FKN8 A0A1A9UFV3 A0A1A9WSM9 A0A0A9XNI8
A0A1E1W6H0 A0A2H1VS46 H9IUK1 A0A212EZJ3 A0A194RHU6 A0A212EZJ4 A0A194Q6F0 I4DKN7 I4DRB7 A0A194QVX4 S4PL71 A0A067RBB7 A0A1B0CUL9 A0A1B6KQM6 A0A1L8DGU4 A0A1L8DGV1 A0A1B6HZF3 A0A1B6GM18 A0A1L8DGR3 A0A336K9F1 A0A1B6BXZ8 A0A336KBA5 A0A0M4ENG0 A0A0Q9XR26 A0A1B6EAQ6 B4L164 A0A0K8TQV2 A0A0Q9WM05 W8ASK5 A0A3B0JPQ5 A0A0P9AKQ5 B4LC30 A0A3B0K245 B3MAG0 W8B455 B4H1W5 A0A034W2B8 A0A0K8WAS3 Q29EE0 A0A0K8TV29 A0A034W6F8 A0A0K8UNG5 A0A0R1E1Z7 A0A0Q5UJ14 B4MLI7 A0A3B0JUS5 B4PG79 B3NH50 Q16VZ2 A0A1W4W603 Q16VZ3 B4IXY9 A0A182GKR0 A0A224XJ77 U4UD40 A0A0P4VJX9 A0A023F5F6 A0A069DSE9 A0A1W4W7P3 J3JV26 A0A084WI87 Q8I0I5 A0A1I8PU18 A0A1I8PU50 V5GRD5 T1P857 Q9VTS4 A0A0L0BZQ3 A0A0A9XQT1 A0A023ES05 A0A1I8N2Q5 A0A1I8PU32 A0A1I8N2Q9 A0A1Y1LSN9 A0A0T6B7Y0 A0A0J9RU39 A0A1I8JT44 Q8IGD6 A0A0A9XKD7 A0A182WR57 A0A182LPR2 Q7Q2Z8 D2A3D8 A0A0J9RTR8 A0A1B0C4U7 A0A0P5W4P6 A0A2P8XSW5 D3TP58 A0A1A9XGF6 A0A1A9ZKN2 A0A1B0FKN8 A0A1A9UFV3 A0A1A9WSM9 A0A0A9XNI8
Pubmed
26354079
22651552
19121390
22118469
23622113
24845553
+ More
17994087 18057021 26369729 24495485 25348373 15632085 17550304 17510324 26483478 23537049 27129103 25474469 26334808 22516182 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 26108605 25401762 24945155 28004739 22936249 26823975 20966253 12364791 14747013 17210077 18362917 19820115 29403074 20353571
17994087 18057021 26369729 24495485 25348373 15632085 17550304 17510324 26483478 23537049 27129103 25474469 26334808 22516182 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 26108605 25401762 24945155 28004739 22936249 26823975 20966253 12364791 14747013 17210077 18362917 19820115 29403074 20353571
EMBL
GDQN01004386
JAT86668.1
NWSH01002871
PCG67408.1
ODYU01011861
SOQ57846.1
+ More
KQ459606 KPI91274.1 AK404915 BAM20372.1 PCG67406.1 GDQN01008478 JAT82576.1 ODYU01004110 SOQ43640.1 BABH01000938 BABH01000939 BABH01000940 BABH01000941 AGBW02011268 OWR46919.1 KQ460205 KPJ16900.1 OWR46918.1 KQ459463 KPJ00580.1 AK401855 BAM18477.1 AK405054 BAM20457.1 KQ461108 KPJ09115.1 GAIX01004205 JAA88355.1 KK852756 KDR17086.1 AJWK01029371 AJWK01029372 GEBQ01031228 GEBQ01026231 GEBQ01002950 GEBQ01001848 JAT08749.1 JAT13746.1 JAT37027.1 JAT38129.1 GFDF01008412 JAV05672.1 GFDF01008510 JAV05574.1 GECU01027649 JAS80057.1 GECZ01009579 GECZ01006280 JAS60190.1 JAS63489.1 GFDF01008463 JAV05621.1 UFQS01000195 UFQT01000195 SSX01121.1 SSX21501.1 GEDC01031162 JAS06136.1 SSX01122.1 SSX21502.1 CP012525 ALC43480.1 CH933809 KRG06567.1 KRG06568.1 GEDC01002305 JAS34993.1 EDW19246.2 GDAI01000861 JAI16742.1 CH940647 KRF85158.1 GAMC01014706 GAMC01014705 JAB91850.1 OUUW01000002 SPP77460.1 CH902618 KPU78448.1 EDW70858.1 SPP77458.1 EDV40211.2 GAMC01014707 JAB91848.1 CH479203 EDW30317.1 GAKP01009261 JAC49691.1 GDHF01033152 GDHF01007000 GDHF01004083 GDHF01000922 JAI19162.1 JAI45314.1 JAI48231.1 JAI51392.1 CH379070 EAL30121.3 GDHF01033992 JAI18322.1 GAKP01009262 JAC49690.1 GDHF01024419 JAI27895.1 CM000159 KRK01713.1 CH954178 KQS43824.1 CH963847 EDW72843.2 SPP77459.1 EDW94243.1 EDV51507.1 CH477579 EAT38721.1 EAT38720.1 CH916366 EDV97532.1 JXUM01070467 JXUM01070468 JXUM01070469 JXUM01070470 GFTR01005322 JAW11104.1 KB632266 ERL90972.1 GDKW01001261 JAI55334.1 GBBI01002339 JAC16373.1 GBGD01002297 JAC86592.1 BT127092 AEE62054.1 ATLV01023923 KE525347 KFB49931.1 AE014296 BT001837 AAN12248.1 AAN12249.1 AAN71594.1 GALX01005643 JAB62823.1 KA644841 AFP59470.1 AY047553 AAF49972.2 AAK77285.1 JRES01001098 KNC25525.1 GBHO01022421 JAG21183.1 GAPW01001595 JAC12003.1 GEZM01047870 GEZM01047869 JAV76714.1 LJIG01009261 KRT83433.1 CM002912 KMY99133.1 KMY99137.1 APCN01001962 BT001830 AAN71585.1 GBHO01022427 GDHC01001711 JAG21177.1 JAQ16918.1 AAAB01008966 EAA12934.4 KQ971338 EFA02299.1 KMY99138.1 JXJN01025657 GDIP01091162 JAM12553.1 PYGN01001401 PSN35098.1 EZ423210 ADD19486.1 CCAG010006132 GBHO01022418 JAG21186.1
KQ459606 KPI91274.1 AK404915 BAM20372.1 PCG67406.1 GDQN01008478 JAT82576.1 ODYU01004110 SOQ43640.1 BABH01000938 BABH01000939 BABH01000940 BABH01000941 AGBW02011268 OWR46919.1 KQ460205 KPJ16900.1 OWR46918.1 KQ459463 KPJ00580.1 AK401855 BAM18477.1 AK405054 BAM20457.1 KQ461108 KPJ09115.1 GAIX01004205 JAA88355.1 KK852756 KDR17086.1 AJWK01029371 AJWK01029372 GEBQ01031228 GEBQ01026231 GEBQ01002950 GEBQ01001848 JAT08749.1 JAT13746.1 JAT37027.1 JAT38129.1 GFDF01008412 JAV05672.1 GFDF01008510 JAV05574.1 GECU01027649 JAS80057.1 GECZ01009579 GECZ01006280 JAS60190.1 JAS63489.1 GFDF01008463 JAV05621.1 UFQS01000195 UFQT01000195 SSX01121.1 SSX21501.1 GEDC01031162 JAS06136.1 SSX01122.1 SSX21502.1 CP012525 ALC43480.1 CH933809 KRG06567.1 KRG06568.1 GEDC01002305 JAS34993.1 EDW19246.2 GDAI01000861 JAI16742.1 CH940647 KRF85158.1 GAMC01014706 GAMC01014705 JAB91850.1 OUUW01000002 SPP77460.1 CH902618 KPU78448.1 EDW70858.1 SPP77458.1 EDV40211.2 GAMC01014707 JAB91848.1 CH479203 EDW30317.1 GAKP01009261 JAC49691.1 GDHF01033152 GDHF01007000 GDHF01004083 GDHF01000922 JAI19162.1 JAI45314.1 JAI48231.1 JAI51392.1 CH379070 EAL30121.3 GDHF01033992 JAI18322.1 GAKP01009262 JAC49690.1 GDHF01024419 JAI27895.1 CM000159 KRK01713.1 CH954178 KQS43824.1 CH963847 EDW72843.2 SPP77459.1 EDW94243.1 EDV51507.1 CH477579 EAT38721.1 EAT38720.1 CH916366 EDV97532.1 JXUM01070467 JXUM01070468 JXUM01070469 JXUM01070470 GFTR01005322 JAW11104.1 KB632266 ERL90972.1 GDKW01001261 JAI55334.1 GBBI01002339 JAC16373.1 GBGD01002297 JAC86592.1 BT127092 AEE62054.1 ATLV01023923 KE525347 KFB49931.1 AE014296 BT001837 AAN12248.1 AAN12249.1 AAN71594.1 GALX01005643 JAB62823.1 KA644841 AFP59470.1 AY047553 AAF49972.2 AAK77285.1 JRES01001098 KNC25525.1 GBHO01022421 JAG21183.1 GAPW01001595 JAC12003.1 GEZM01047870 GEZM01047869 JAV76714.1 LJIG01009261 KRT83433.1 CM002912 KMY99133.1 KMY99137.1 APCN01001962 BT001830 AAN71585.1 GBHO01022427 GDHC01001711 JAG21177.1 JAQ16918.1 AAAB01008966 EAA12934.4 KQ971338 EFA02299.1 KMY99138.1 JXJN01025657 GDIP01091162 JAM12553.1 PYGN01001401 PSN35098.1 EZ423210 ADD19486.1 CCAG010006132 GBHO01022418 JAG21186.1
Proteomes
UP000218220
UP000053268
UP000005204
UP000007151
UP000053240
UP000027135
+ More
UP000092461 UP000092553 UP000009192 UP000008792 UP000268350 UP000007801 UP000008744 UP000001819 UP000002282 UP000008711 UP000007798 UP000008820 UP000192221 UP000001070 UP000069940 UP000030742 UP000192223 UP000030765 UP000000803 UP000095300 UP000095301 UP000037069 UP000075840 UP000076407 UP000075882 UP000007062 UP000007266 UP000092460 UP000245037 UP000092443 UP000092445 UP000092444 UP000078200 UP000091820
UP000092461 UP000092553 UP000009192 UP000008792 UP000268350 UP000007801 UP000008744 UP000001819 UP000002282 UP000008711 UP000007798 UP000008820 UP000192221 UP000001070 UP000069940 UP000030742 UP000192223 UP000030765 UP000000803 UP000095300 UP000095301 UP000037069 UP000075840 UP000076407 UP000075882 UP000007062 UP000007266 UP000092460 UP000245037 UP000092443 UP000092445 UP000092444 UP000078200 UP000091820
Pfam
PF04857 CAF1
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
A0A1E1WI84
A0A2A4J6V5
A0A2H1WY06
A0A194PEZ7
I4DR32
A0A2A4J6P5
+ More
A0A1E1W6H0 A0A2H1VS46 H9IUK1 A0A212EZJ3 A0A194RHU6 A0A212EZJ4 A0A194Q6F0 I4DKN7 I4DRB7 A0A194QVX4 S4PL71 A0A067RBB7 A0A1B0CUL9 A0A1B6KQM6 A0A1L8DGU4 A0A1L8DGV1 A0A1B6HZF3 A0A1B6GM18 A0A1L8DGR3 A0A336K9F1 A0A1B6BXZ8 A0A336KBA5 A0A0M4ENG0 A0A0Q9XR26 A0A1B6EAQ6 B4L164 A0A0K8TQV2 A0A0Q9WM05 W8ASK5 A0A3B0JPQ5 A0A0P9AKQ5 B4LC30 A0A3B0K245 B3MAG0 W8B455 B4H1W5 A0A034W2B8 A0A0K8WAS3 Q29EE0 A0A0K8TV29 A0A034W6F8 A0A0K8UNG5 A0A0R1E1Z7 A0A0Q5UJ14 B4MLI7 A0A3B0JUS5 B4PG79 B3NH50 Q16VZ2 A0A1W4W603 Q16VZ3 B4IXY9 A0A182GKR0 A0A224XJ77 U4UD40 A0A0P4VJX9 A0A023F5F6 A0A069DSE9 A0A1W4W7P3 J3JV26 A0A084WI87 Q8I0I5 A0A1I8PU18 A0A1I8PU50 V5GRD5 T1P857 Q9VTS4 A0A0L0BZQ3 A0A0A9XQT1 A0A023ES05 A0A1I8N2Q5 A0A1I8PU32 A0A1I8N2Q9 A0A1Y1LSN9 A0A0T6B7Y0 A0A0J9RU39 A0A1I8JT44 Q8IGD6 A0A0A9XKD7 A0A182WR57 A0A182LPR2 Q7Q2Z8 D2A3D8 A0A0J9RTR8 A0A1B0C4U7 A0A0P5W4P6 A0A2P8XSW5 D3TP58 A0A1A9XGF6 A0A1A9ZKN2 A0A1B0FKN8 A0A1A9UFV3 A0A1A9WSM9 A0A0A9XNI8
A0A1E1W6H0 A0A2H1VS46 H9IUK1 A0A212EZJ3 A0A194RHU6 A0A212EZJ4 A0A194Q6F0 I4DKN7 I4DRB7 A0A194QVX4 S4PL71 A0A067RBB7 A0A1B0CUL9 A0A1B6KQM6 A0A1L8DGU4 A0A1L8DGV1 A0A1B6HZF3 A0A1B6GM18 A0A1L8DGR3 A0A336K9F1 A0A1B6BXZ8 A0A336KBA5 A0A0M4ENG0 A0A0Q9XR26 A0A1B6EAQ6 B4L164 A0A0K8TQV2 A0A0Q9WM05 W8ASK5 A0A3B0JPQ5 A0A0P9AKQ5 B4LC30 A0A3B0K245 B3MAG0 W8B455 B4H1W5 A0A034W2B8 A0A0K8WAS3 Q29EE0 A0A0K8TV29 A0A034W6F8 A0A0K8UNG5 A0A0R1E1Z7 A0A0Q5UJ14 B4MLI7 A0A3B0JUS5 B4PG79 B3NH50 Q16VZ2 A0A1W4W603 Q16VZ3 B4IXY9 A0A182GKR0 A0A224XJ77 U4UD40 A0A0P4VJX9 A0A023F5F6 A0A069DSE9 A0A1W4W7P3 J3JV26 A0A084WI87 Q8I0I5 A0A1I8PU18 A0A1I8PU50 V5GRD5 T1P857 Q9VTS4 A0A0L0BZQ3 A0A0A9XQT1 A0A023ES05 A0A1I8N2Q5 A0A1I8PU32 A0A1I8N2Q9 A0A1Y1LSN9 A0A0T6B7Y0 A0A0J9RU39 A0A1I8JT44 Q8IGD6 A0A0A9XKD7 A0A182WR57 A0A182LPR2 Q7Q2Z8 D2A3D8 A0A0J9RTR8 A0A1B0C4U7 A0A0P5W4P6 A0A2P8XSW5 D3TP58 A0A1A9XGF6 A0A1A9ZKN2 A0A1B0FKN8 A0A1A9UFV3 A0A1A9WSM9 A0A0A9XNI8
PDB
4GMJ
E-value=5.24992e-110,
Score=1015
Ontologies
GO
PANTHER
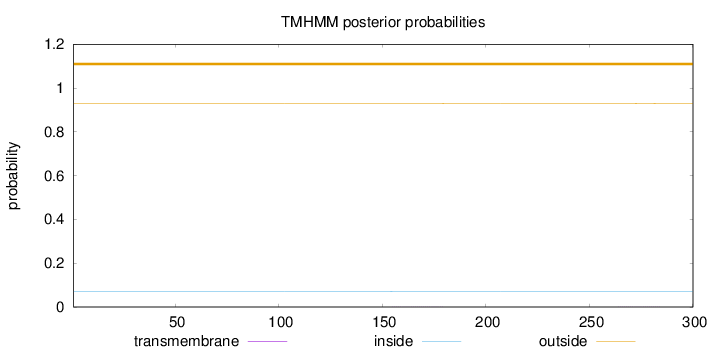
Topology
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01344
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.07038
outside
1 - 300
Population Genetic Test Statistics
Pi
267.114304
Theta
208.750235
Tajima's D
0.88264
CLR
0
CSRT
0.635718214089296
Interpretation
Uncertain
