Gene
KWMTBOMO07824
Annotation
PREDICTED:_uncharacterized_protein_LOC106131292_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.385 Nuclear Reliability : 1.149
Sequence
CDS
ATGGAAAGTCATACAGTTGCTGTATATCATTTTTTGGATTGGGAGTCTGCAATGAATCGGCTTAGTGAAAAAGCTGTGCTCCTCCTGTCAAAAAAATATACAGATTATGTACCATCTGAAAATTACAAAAAACAGTACAATATTAGTGAAAAAATGATGCTAGATATTATGAAAGTTTTGGGAGATTGTCGAGGCGGCCAAAATTATGTCATAGCTGAACATATTTTAACCCACCATCAAAGAGGAGATATAATCATTTGCAATGATCACAATGGAAAGCCTATGCCAGTCAAAGAAGCATTTAAAAATGATACATTTGGGTATATTCGGAAGCCTCCTGATGACAATGATTGGATTGTACTTATAATAGGTGGAAAAAATTCTCTAATTCAGAACTGTAATGTACCTACAGGACAGTTTCAGACAAAAATTCGAGAACTTGCAGCTTTAGGCTACACTCCAATATTGGTGCCATGGAAAACATATTCACAACTAGATAATTTTGAAGAGAAAGAAAGTTATCTTTGTAGCTTAATCGAAAACAGATCAAGGAAAAATCATAGTTGTACTGATACTGTATTTAATAGTCTTCATAGTCATAATTCTTAG
Protein
MESHTVAVYHFLDWESAMNRLSEKAVLLLSKKYTDYVPSENYKKQYNISEKMMLDIMKVLGDCRGGQNYVIAEHILTHHQRGDIIICNDHNGKPMPVKEAFKNDTFGYIRKPPDDNDWIVLIIGGKNSLIQNCNVPTGQFQTKIRELAALGYTPILVPWKTYSQLDNFEEKESYLCSLIENRSRKNHSCTDTVFNSLHSHNS
Summary
Uniprot
A0A2A4J7J0
A0A2H1WMD7
A0A194RGF3
S4PF89
A0A194PDE8
A0A212ESI0
+ More
A0A0L7LD22 A0A2J7Q150 A0A067R5M1 A0A1B6EGB2 A0A1W4WXS9 A0A1W4WXV2 A0A1W4X8M0 A0A139WGR7 U4UQ97 N6TTF4 A0A0P4WAU2 A0A1Y1NF02 A0A1Y1NF06 A0A1Y1NF33 B4H0E0 A0A0P4VWT8 A0A3B0KV64 B3MSA4 Q29H39 B4M1Q8 A0A1B0FIJ7 A0A224X9B2 A0A1I8PXS6 Q16ZQ8 B4PXX9 A0A1S4FZM3 B3NW17 A0A0K8TZK4 A0A034V7E9 B4JKN8 Q16IK7 A0A1I8MC20 B4R7L1 Q86CW0 Q9W2T3 A0A1W4VJA8 A0A1L8DJG3 B4IDS1 A0A1J1IXC0 A0A1A9ULB0 A0A0A1XP34 A0A1B0AE97 W8BT31 W8CC49 A0A195D1P1 B4MT66 A0A1B0APJ9 A0A0C9S074 Q86CW2 A0A0M3QZ50 A0A1A9YA26 A0A182F8A5 A0A182KFJ5 A0A026WDN2 B4L8T0 A0A182MK28 A0A182IIX8 A0A182PBJ0 F4WE82 A0A151J502 A0A0K8SNP8 A0A1A9WNF9 A0A182Q0T6 A0A1Q3G2C0 A0A158P187 A0A182N193 A0A336M7Y5 A0A336M3W2 A0A146LY28 A0A182XXT7 A0A0L0CKC9
A0A0L7LD22 A0A2J7Q150 A0A067R5M1 A0A1B6EGB2 A0A1W4WXS9 A0A1W4WXV2 A0A1W4X8M0 A0A139WGR7 U4UQ97 N6TTF4 A0A0P4WAU2 A0A1Y1NF02 A0A1Y1NF06 A0A1Y1NF33 B4H0E0 A0A0P4VWT8 A0A3B0KV64 B3MSA4 Q29H39 B4M1Q8 A0A1B0FIJ7 A0A224X9B2 A0A1I8PXS6 Q16ZQ8 B4PXX9 A0A1S4FZM3 B3NW17 A0A0K8TZK4 A0A034V7E9 B4JKN8 Q16IK7 A0A1I8MC20 B4R7L1 Q86CW0 Q9W2T3 A0A1W4VJA8 A0A1L8DJG3 B4IDS1 A0A1J1IXC0 A0A1A9ULB0 A0A0A1XP34 A0A1B0AE97 W8BT31 W8CC49 A0A195D1P1 B4MT66 A0A1B0APJ9 A0A0C9S074 Q86CW2 A0A0M3QZ50 A0A1A9YA26 A0A182F8A5 A0A182KFJ5 A0A026WDN2 B4L8T0 A0A182MK28 A0A182IIX8 A0A182PBJ0 F4WE82 A0A151J502 A0A0K8SNP8 A0A1A9WNF9 A0A182Q0T6 A0A1Q3G2C0 A0A158P187 A0A182N193 A0A336M7Y5 A0A336M3W2 A0A146LY28 A0A182XXT7 A0A0L0CKC9
Pubmed
26354079
23622113
22118469
26227816
24845553
18362917
+ More
19820115 23537049 28004739 17994087 27129103 15632085 17510324 17550304 25348373 25315136 12695567 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 24508170 30249741 21719571 21347285 26823975 25244985 26108605
19820115 23537049 28004739 17994087 27129103 15632085 17510324 17550304 25348373 25315136 12695567 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 24495485 24508170 30249741 21719571 21347285 26823975 25244985 26108605
EMBL
NWSH01002871
PCG67410.1
ODYU01009651
SOQ54229.1
KQ460205
KPJ16898.1
+ More
GAIX01004147 JAA88413.1 KQ459606 KPI91272.1 AGBW02012794 OWR44429.1 JTDY01001600 KOB73408.1 NEVH01019960 PNF22310.1 KK852945 KDR13439.1 GEDC01000316 JAS36982.1 KQ971344 KYB27089.1 KB632413 ERL95272.1 APGK01055293 KB741261 ENN71646.1 GDRN01058296 GDRN01058294 JAI65611.1 GEZM01003867 JAV96513.1 GEZM01003866 JAV96514.1 GEZM01003868 JAV96512.1 CH479200 EDW29735.1 GDKW01002538 JAI54057.1 OUUW01000019 SPP89241.1 CH902622 EDV34659.1 CH379064 EAL31920.1 CH940651 EDW65612.1 CCAG010023071 GFTR01007500 JAW08926.1 CH477485 EAT40164.1 CM000162 EDX02951.1 CH954180 EDV46150.1 GDHF01032608 GDHF01015780 JAI19706.1 JAI36534.1 GAKP01019761 GAKP01019759 JAC39191.1 CH916370 EDW00141.1 CH478072 EAT34100.1 CM000366 EDX17570.1 AY260056 AAO85035.1 AE014298 AY071525 AY260057 AAF46606.1 AAL49147.1 AAO85037.1 GFDF01007584 JAV06500.1 CH480830 EDW45729.1 CVRI01000062 CRL04212.1 GBXI01001585 JAD12707.1 GAMC01004218 JAC02338.1 GAMC01004221 JAC02335.1 KQ976956 KYN06823.1 CH963851 EDW75305.2 JXJN01001458 GBYB01013876 JAG83643.1 AY260055 AAO85033.1 CP012528 ALC48813.1 KK107273 QOIP01000012 EZA53791.1 RLU16334.1 CH933815 EDW08055.2 AXCM01000597 GL888102 EGI67520.1 KQ980071 KYN17853.1 GBRD01010893 JAG54931.1 AXCN02001237 GFDL01001078 JAV33967.1 ADTU01000071 UFQS01000524 UFQT01000524 SSX04590.1 SSX24953.1 SSX04591.1 SSX24954.1 GDHC01006101 JAQ12528.1 JRES01000290 KNC32715.1
GAIX01004147 JAA88413.1 KQ459606 KPI91272.1 AGBW02012794 OWR44429.1 JTDY01001600 KOB73408.1 NEVH01019960 PNF22310.1 KK852945 KDR13439.1 GEDC01000316 JAS36982.1 KQ971344 KYB27089.1 KB632413 ERL95272.1 APGK01055293 KB741261 ENN71646.1 GDRN01058296 GDRN01058294 JAI65611.1 GEZM01003867 JAV96513.1 GEZM01003866 JAV96514.1 GEZM01003868 JAV96512.1 CH479200 EDW29735.1 GDKW01002538 JAI54057.1 OUUW01000019 SPP89241.1 CH902622 EDV34659.1 CH379064 EAL31920.1 CH940651 EDW65612.1 CCAG010023071 GFTR01007500 JAW08926.1 CH477485 EAT40164.1 CM000162 EDX02951.1 CH954180 EDV46150.1 GDHF01032608 GDHF01015780 JAI19706.1 JAI36534.1 GAKP01019761 GAKP01019759 JAC39191.1 CH916370 EDW00141.1 CH478072 EAT34100.1 CM000366 EDX17570.1 AY260056 AAO85035.1 AE014298 AY071525 AY260057 AAF46606.1 AAL49147.1 AAO85037.1 GFDF01007584 JAV06500.1 CH480830 EDW45729.1 CVRI01000062 CRL04212.1 GBXI01001585 JAD12707.1 GAMC01004218 JAC02338.1 GAMC01004221 JAC02335.1 KQ976956 KYN06823.1 CH963851 EDW75305.2 JXJN01001458 GBYB01013876 JAG83643.1 AY260055 AAO85033.1 CP012528 ALC48813.1 KK107273 QOIP01000012 EZA53791.1 RLU16334.1 CH933815 EDW08055.2 AXCM01000597 GL888102 EGI67520.1 KQ980071 KYN17853.1 GBRD01010893 JAG54931.1 AXCN02001237 GFDL01001078 JAV33967.1 ADTU01000071 UFQS01000524 UFQT01000524 SSX04590.1 SSX24953.1 SSX04591.1 SSX24954.1 GDHC01006101 JAQ12528.1 JRES01000290 KNC32715.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000235965
+ More
UP000027135 UP000192223 UP000007266 UP000030742 UP000019118 UP000008744 UP000268350 UP000007801 UP000001819 UP000008792 UP000092444 UP000095300 UP000008820 UP000002282 UP000008711 UP000001070 UP000095301 UP000000304 UP000000803 UP000192221 UP000001292 UP000183832 UP000078200 UP000092445 UP000078542 UP000007798 UP000092460 UP000092553 UP000092443 UP000069272 UP000075881 UP000053097 UP000279307 UP000009192 UP000075883 UP000075880 UP000075885 UP000007755 UP000078492 UP000091820 UP000075886 UP000005205 UP000075884 UP000076408 UP000037069
UP000027135 UP000192223 UP000007266 UP000030742 UP000019118 UP000008744 UP000268350 UP000007801 UP000001819 UP000008792 UP000092444 UP000095300 UP000008820 UP000002282 UP000008711 UP000001070 UP000095301 UP000000304 UP000000803 UP000192221 UP000001292 UP000183832 UP000078200 UP000092445 UP000078542 UP000007798 UP000092460 UP000092553 UP000092443 UP000069272 UP000075881 UP000053097 UP000279307 UP000009192 UP000075883 UP000075880 UP000075885 UP000007755 UP000078492 UP000091820 UP000075886 UP000005205 UP000075884 UP000076408 UP000037069
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A2A4J7J0
A0A2H1WMD7
A0A194RGF3
S4PF89
A0A194PDE8
A0A212ESI0
+ More
A0A0L7LD22 A0A2J7Q150 A0A067R5M1 A0A1B6EGB2 A0A1W4WXS9 A0A1W4WXV2 A0A1W4X8M0 A0A139WGR7 U4UQ97 N6TTF4 A0A0P4WAU2 A0A1Y1NF02 A0A1Y1NF06 A0A1Y1NF33 B4H0E0 A0A0P4VWT8 A0A3B0KV64 B3MSA4 Q29H39 B4M1Q8 A0A1B0FIJ7 A0A224X9B2 A0A1I8PXS6 Q16ZQ8 B4PXX9 A0A1S4FZM3 B3NW17 A0A0K8TZK4 A0A034V7E9 B4JKN8 Q16IK7 A0A1I8MC20 B4R7L1 Q86CW0 Q9W2T3 A0A1W4VJA8 A0A1L8DJG3 B4IDS1 A0A1J1IXC0 A0A1A9ULB0 A0A0A1XP34 A0A1B0AE97 W8BT31 W8CC49 A0A195D1P1 B4MT66 A0A1B0APJ9 A0A0C9S074 Q86CW2 A0A0M3QZ50 A0A1A9YA26 A0A182F8A5 A0A182KFJ5 A0A026WDN2 B4L8T0 A0A182MK28 A0A182IIX8 A0A182PBJ0 F4WE82 A0A151J502 A0A0K8SNP8 A0A1A9WNF9 A0A182Q0T6 A0A1Q3G2C0 A0A158P187 A0A182N193 A0A336M7Y5 A0A336M3W2 A0A146LY28 A0A182XXT7 A0A0L0CKC9
A0A0L7LD22 A0A2J7Q150 A0A067R5M1 A0A1B6EGB2 A0A1W4WXS9 A0A1W4WXV2 A0A1W4X8M0 A0A139WGR7 U4UQ97 N6TTF4 A0A0P4WAU2 A0A1Y1NF02 A0A1Y1NF06 A0A1Y1NF33 B4H0E0 A0A0P4VWT8 A0A3B0KV64 B3MSA4 Q29H39 B4M1Q8 A0A1B0FIJ7 A0A224X9B2 A0A1I8PXS6 Q16ZQ8 B4PXX9 A0A1S4FZM3 B3NW17 A0A0K8TZK4 A0A034V7E9 B4JKN8 Q16IK7 A0A1I8MC20 B4R7L1 Q86CW0 Q9W2T3 A0A1W4VJA8 A0A1L8DJG3 B4IDS1 A0A1J1IXC0 A0A1A9ULB0 A0A0A1XP34 A0A1B0AE97 W8BT31 W8CC49 A0A195D1P1 B4MT66 A0A1B0APJ9 A0A0C9S074 Q86CW2 A0A0M3QZ50 A0A1A9YA26 A0A182F8A5 A0A182KFJ5 A0A026WDN2 B4L8T0 A0A182MK28 A0A182IIX8 A0A182PBJ0 F4WE82 A0A151J502 A0A0K8SNP8 A0A1A9WNF9 A0A182Q0T6 A0A1Q3G2C0 A0A158P187 A0A182N193 A0A336M7Y5 A0A336M3W2 A0A146LY28 A0A182XXT7 A0A0L0CKC9
Ontologies
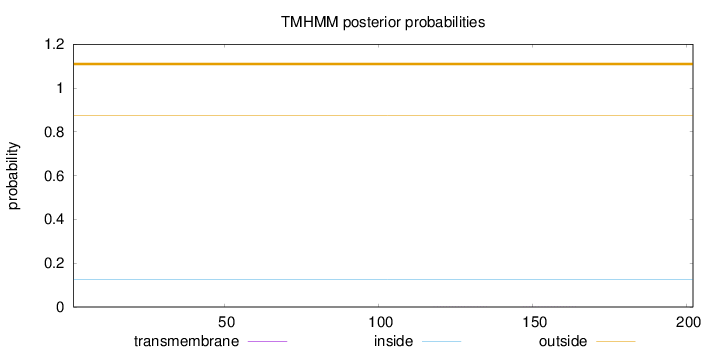
Topology
Length:
202
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00691
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.12663
outside
1 - 202
Population Genetic Test Statistics
Pi
129.957509
Theta
141.519751
Tajima's D
-0.394449
CLR
136.393794
CSRT
0.264336783160842
Interpretation
Uncertain
