Gene
KWMTBOMO07820 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001218
Annotation
PREDICTED:_97_kDa_heat_shock_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.592 Nuclear Reliability : 2.086
Sequence
CDS
ATGGCTGCTATGTCCGTCATTGGAATTGATTTTGGTAATGAATCCTGCTATATTGCTGTGGCTAAAGCAGGTGGGATAGAGACAATAGCCAATGATTACAGTCTTCGAGGGACACCATCATGTGTTGCTTTTTCACAAAAGAATCGAATATTGGGTGTGGCAGCCAAGAACCAAATGGTGACCAATATGAAGAATACTGTATTTGGTTTCAAACGACTTCTGGGAAGAAAATTTTCAGACCCTCTTGTTCAAAAAGAATTAAAACATTTCCCTTTCAAGGTTGAACAGAGGTCTGATGGCGGTATTGGAATAAGAGTCAACTACCTCAATGAAGACAATGTCTTTACCCCTGAGCAGATAACAGCCATGTTGTTGACAAAACTAAAAGATGTTGCAGCAACAGCTTTACAGACTCAAATCAACGACTGTGTGATATCTGTTCCCAGCTATTTCACGAATGCTGAAAGAAATGCTCTTTTGAATGCCGCTGGTATTGCTGGTCTTAATGTATTGCGCCTCATGAACGAGACAACTGCGACAGCTTTAACTTATGGAATTTATAAACAAGATCTTCCTGGTGCTGAAGAGAAACCACGAAATGTTGTCTTTGTCGACTTTGGTCACAGTTCTTTACAGGTTTCAGCTTGTGCTTTCAATAAGGGTAAGCTGCGTATTCTCGCTACTGCAACCGATGCTCAGTGCGGTGGACGAGACATTGACATGGCTTTAGCTGATTACTTCACTCAAGATTTTGTGACAAGATATAAATTAGACGCCAGAAAAAATCAAAGGGCTTATCTTCGTCTTCTGCAAGAAGTTGAGAAATTGAAAAAACAAATGTCTGCTAACAGTACTCGTTTACCACTTAACATTGAGTGCTTTATGGAAGAACGAGATGTATCTGGAGATATGCAGCGTTCTCAGATGGAACAAATATGTGCTGAAACATTTAACCGCGTTGAAAGAACCTTAAGAGCTATTCTACATAATGCAAAACTAAGGTCTGAAGACATCCACTCTGTTGAAATTGTTGGTGGATCCACCAGAATCCCTGCAGTTAAGAGTTTGATTGAACAAGCGTTTGGCAAGCATGCTTCCACAACCTTGAATCAGGACGAGGCGGTGGCTCGCGGATGCGCGCTACAGTGCGCCATGCTGAGCCCGGCCGTACGCGTGCGTGAGTTCAGCATCACCGACGTGCAGCCCTACGCCGTGCGCCTCGCCTGGGACGCCGCCAGAAACGAGGACGGTGATATGGAGGTATTCCCTGCTTTTCACGCTGCACCATTTTCAAAAATGTTAACATTCTACAGGAAAGAGCCATTTTCTGTCAGTGCCTATTATTCTGATCAAGTACCTTATCCGGACACATTTATTGGCCAGTGGCAAATCAAAGATGTCCAGCCCACACCTGAAGGGGAATCGCAAAAAGTGAAATTGAAAGTGAGGGTGAACATTCATGGCATCATAACAGTCGCTTCGGCTTCACTTGTTGAAAAGAAGCAGGACACTACACAGAATGAGAACATAGAAATGGAAAATAGTAATGAGAATGCTCAAAATCAGGATACACCTATGGAAACGAATGGAGGAAATCAAGACCAGCAACAACAGAATGGTCCTGAGAACAACCAAGAGGAGAGTGATGATAAAAAGGATAAATCCAAGGATAAGTCAAAGAAGGTTCTTGTGAAAACTGTGGAGCTGCCCATTGAAGCACAAACACATGGTCTATCAGTGCATGAACTAAATGGATATGTTGAACAAGAGGGCAAAATGCAAGCTCAAGATCGGCAAGAGAAAGAAAGAGCAGATGCTCGTAATGCTCTTGAAGAATATGTTTACGAATTGCGAGGGAAACTGTCTGAAGGCGAAACCCTGCATGATTTTGTTGCAGAAGATCAACGTAATAAGCTTGTCAATCATCTTGATTCTGTTGAACAATGGCTCTATGATGAAGGTGAAGACCAAAATAGACAGGTGTACAGTGATAAGCTGAGCGAGTTGAAAACTGAAGGCGAGCCGATCAAACAAAGGCGTCTTGAGTTTGAGCTACGTCCTGGGGCTTTAGATGAGTTTGCTTTATCCATCCAACTCACCAACAAAGCTGTTGATTTATACAAAGCTGGTGACGCTAAATTTTCACACTTGGCGGAAACAGAGGTTCAGAAAGTGATAGACGCATCTAAAAATGCCCTAACTTGGCTCGAGACTGCAAGGCAAGCGTTAGCACACACGCCACGTCATCAGCAACCATCGCACACTACTCATCAAATCCGTCAAGAACGTCAGAATTTTGAGAACCTCGTGCATCCAATTTTGAATAAAGTTAAACCAAAAGAGAAAACACCACCTCCTTCCAATCAGACTGCGGGAGACGGACAGGCTGATGGTAACAAGGATGCGCCTTCGACTCAATCTCAAGAGCAAATGGATGTTGAATAA
Protein
MAAMSVIGIDFGNESCYIAVAKAGGIETIANDYSLRGTPSCVAFSQKNRILGVAAKNQMVTNMKNTVFGFKRLLGRKFSDPLVQKELKHFPFKVEQRSDGGIGIRVNYLNEDNVFTPEQITAMLLTKLKDVAATALQTQINDCVISVPSYFTNAERNALLNAAGIAGLNVLRLMNETTATALTYGIYKQDLPGAEEKPRNVVFVDFGHSSLQVSACAFNKGKLRILATATDAQCGGRDIDMALADYFTQDFVTRYKLDARKNQRAYLRLLQEVEKLKKQMSANSTRLPLNIECFMEERDVSGDMQRSQMEQICAETFNRVERTLRAILHNAKLRSEDIHSVEIVGGSTRIPAVKSLIEQAFGKHASTTLNQDEAVARGCALQCAMLSPAVRVREFSITDVQPYAVRLAWDAARNEDGDMEVFPAFHAAPFSKMLTFYRKEPFSVSAYYSDQVPYPDTFIGQWQIKDVQPTPEGESQKVKLKVRVNIHGIITVASASLVEKKQDTTQNENIEMENSNENAQNQDTPMETNGGNQDQQQQNGPENNQEESDDKKDKSKDKSKKVLVKTVELPIEAQTHGLSVHELNGYVEQEGKMQAQDRQEKERADARNALEEYVYELRGKLSEGETLHDFVAEDQRNKLVNHLDSVEQWLYDEGEDQNRQVYSDKLSELKTEGEPIKQRRLEFELRPGALDEFALSIQLTNKAVDLYKAGDAKFSHLAETEVQKVIDASKNALTWLETARQALAHTPRHQQPSHTTHQIRQERQNFENLVHPILNKVKPKEKTPPPSNQTAGDGQADGNKDAPSTQSQEQMDVE
Summary
Similarity
Belongs to the heat shock protein 70 family.
Uniprot
H9IVD8
A0A2H1VDU5
A0A2A4IWH0
A0A2A4IX17
A0A194RH35
A0A2A4IVL4
+ More
S4PX99 A0A2J7QEY4 A0A1B6C8J9 A0A1B6FM09 A0A1B6GU52 A0A1Y1JTX9 A0A1Y1JQX1 A0A067R776 A0A346THM8 A0A224X7G5 D2A3E8 A0A023F9S1 V9IBC4 U4U4I6 N6UBE3 A0A195E535 A0A195F4P4 R4G605 E2B7Z4 A0A0A9XY29 A0A146L8M9 A0A0K8TST6 A0A0C9R4T8 A0A1B6FPX4 U5EM97 A0A1B6F6H7 A0A1I9WLB6 A0A336MFX0 A0A336M9B2 F2EJC0 J9JVZ3 A0A2S2QMS8 J9HSH6 A0A1Q3FWD8 A0A023EXV4 A0A2H8TT55 A0A2S2PFX9 A0A0A1WZ41 B4KV58 A0A034VZY9 A0A1L8DUC1 A0A1L8DUD5 W8BDM7 A0A1W4U6M6 A0A1W4UK43 A0A0K8U2L3 T1DGM2 A0A0J7KYH4 A0A2M3Z133 A0A0E3T149 A0A0E3T267 A0A1I8MN87 A0A0E3T0V0 A0A2M4A3X9 T1PGF6 A0A182YE12 T1D3U1 A0A0P4WCY3 A0A084VJV5 A0A1I8P985 A0A1I8P9G3 A0A182IL77 A0A0Q9WP46 Q7Q7J4 W5MXU8 A0A3Q0R6W2 A0A3B4ERF4 A0A3B5A539 A0A3Q2WA53 A0A3Q4H4C0 A0A3Q1HJZ1 A0A3P8NZ65 A0A3P9C0W6 A0A3B4XMS6 A0A3B4EPJ4 A0A3B4TQN4 A0A3Q2ZY40 A0A3Q2UTM5 H3CU62 A0A3B4AY19 A0A3B4XHI4 A0A3B4D9D0 A0A3Q3LS94 I3K531 A0A2L1DG12 A0A3Q1GC59 A0A3B5AES1
S4PX99 A0A2J7QEY4 A0A1B6C8J9 A0A1B6FM09 A0A1B6GU52 A0A1Y1JTX9 A0A1Y1JQX1 A0A067R776 A0A346THM8 A0A224X7G5 D2A3E8 A0A023F9S1 V9IBC4 U4U4I6 N6UBE3 A0A195E535 A0A195F4P4 R4G605 E2B7Z4 A0A0A9XY29 A0A146L8M9 A0A0K8TST6 A0A0C9R4T8 A0A1B6FPX4 U5EM97 A0A1B6F6H7 A0A1I9WLB6 A0A336MFX0 A0A336M9B2 F2EJC0 J9JVZ3 A0A2S2QMS8 J9HSH6 A0A1Q3FWD8 A0A023EXV4 A0A2H8TT55 A0A2S2PFX9 A0A0A1WZ41 B4KV58 A0A034VZY9 A0A1L8DUC1 A0A1L8DUD5 W8BDM7 A0A1W4U6M6 A0A1W4UK43 A0A0K8U2L3 T1DGM2 A0A0J7KYH4 A0A2M3Z133 A0A0E3T149 A0A0E3T267 A0A1I8MN87 A0A0E3T0V0 A0A2M4A3X9 T1PGF6 A0A182YE12 T1D3U1 A0A0P4WCY3 A0A084VJV5 A0A1I8P985 A0A1I8P9G3 A0A182IL77 A0A0Q9WP46 Q7Q7J4 W5MXU8 A0A3Q0R6W2 A0A3B4ERF4 A0A3B5A539 A0A3Q2WA53 A0A3Q4H4C0 A0A3Q1HJZ1 A0A3P8NZ65 A0A3P9C0W6 A0A3B4XMS6 A0A3B4EPJ4 A0A3B4TQN4 A0A3Q2ZY40 A0A3Q2UTM5 H3CU62 A0A3B4AY19 A0A3B4XHI4 A0A3B4D9D0 A0A3Q3LS94 I3K531 A0A2L1DG12 A0A3Q1GC59 A0A3B5AES1
Pubmed
EMBL
BABH01000949
ODYU01001837
SOQ38582.1
NWSH01005941
PCG63826.1
PCG63824.1
+ More
KQ460205 KPJ16892.1 PCG63825.1 GAIX01005408 JAA87152.1 NEVH01015305 PNF27137.1 GEDC01027649 JAS09649.1 GECZ01018560 JAS51209.1 GECZ01003896 JAS65873.1 GEZM01103043 JAV51588.1 GEZM01103042 JAV51589.1 KK852655 KDR19282.1 MG265905 AXU24961.1 GFTR01008090 JAW08336.1 KQ971338 EFA02305.1 GBBI01001039 JAC17673.1 JR038766 AEY58400.1 KB631708 ERL85506.1 APGK01030182 APGK01030183 APGK01030184 APGK01030185 APGK01030186 KB740761 ENN79035.1 KQ979608 KYN20273.1 KQ981836 KYN35049.1 GAHY01000060 JAA77450.1 GL446268 EFN88179.1 GBHO01037698 GBHO01021439 GBRD01009141 JAG05906.1 JAG22165.1 JAG56680.1 GDHC01013855 JAQ04774.1 GDAI01000412 JAI17191.1 GBYB01011304 JAG81071.1 GECZ01017568 JAS52201.1 GANO01001126 JAB58745.1 GECZ01023917 JAS45852.1 KU932300 APA33936.1 UFQS01000524 UFQT01000524 SSX04605.1 SSX24968.1 SSX04604.1 SSX24967.1 AK376247 BAK07442.1 ABLF02017872 GGMS01009821 MBY79024.1 CH477328 EJY57568.1 GFDL01003148 JAV31897.1 GAPW01000239 JAC13359.1 GFXV01005608 MBW17413.1 GGMR01015703 MBY28322.1 GBXI01010698 GBXI01010361 JAD03594.1 JAD03931.1 CH933809 EDW19398.1 GAKP01011308 GAKP01011306 GAKP01011305 JAC47647.1 GFDF01004042 JAV10042.1 GFDF01004043 JAV10041.1 GAMC01009758 GAMC01009757 JAB96797.1 GDHF01031724 GDHF01026089 GDHF01018165 GDHF01013860 GDHF01002052 JAI20590.1 JAI26225.1 JAI34149.1 JAI38454.1 JAI50262.1 GALA01000137 JAA94715.1 LBMM01002047 KMQ95354.1 GGFM01001479 MBW22230.1 KR058818 AKB96223.1 KR058819 AKB96224.1 KR058820 AKB96225.1 GGFK01002134 MBW35455.1 KA646958 AFP61587.1 GALA01001243 JAA93609.1 GDRN01062059 JAI65116.1 ATLV01013899 ATLV01013900 KE524908 KFB38249.1 CH963847 KRF97623.1 AAAB01008957 EAA10674.3 AHAT01001545 AERX01036628 AERX01036629 KY753873 AVC68725.1
KQ460205 KPJ16892.1 PCG63825.1 GAIX01005408 JAA87152.1 NEVH01015305 PNF27137.1 GEDC01027649 JAS09649.1 GECZ01018560 JAS51209.1 GECZ01003896 JAS65873.1 GEZM01103043 JAV51588.1 GEZM01103042 JAV51589.1 KK852655 KDR19282.1 MG265905 AXU24961.1 GFTR01008090 JAW08336.1 KQ971338 EFA02305.1 GBBI01001039 JAC17673.1 JR038766 AEY58400.1 KB631708 ERL85506.1 APGK01030182 APGK01030183 APGK01030184 APGK01030185 APGK01030186 KB740761 ENN79035.1 KQ979608 KYN20273.1 KQ981836 KYN35049.1 GAHY01000060 JAA77450.1 GL446268 EFN88179.1 GBHO01037698 GBHO01021439 GBRD01009141 JAG05906.1 JAG22165.1 JAG56680.1 GDHC01013855 JAQ04774.1 GDAI01000412 JAI17191.1 GBYB01011304 JAG81071.1 GECZ01017568 JAS52201.1 GANO01001126 JAB58745.1 GECZ01023917 JAS45852.1 KU932300 APA33936.1 UFQS01000524 UFQT01000524 SSX04605.1 SSX24968.1 SSX04604.1 SSX24967.1 AK376247 BAK07442.1 ABLF02017872 GGMS01009821 MBY79024.1 CH477328 EJY57568.1 GFDL01003148 JAV31897.1 GAPW01000239 JAC13359.1 GFXV01005608 MBW17413.1 GGMR01015703 MBY28322.1 GBXI01010698 GBXI01010361 JAD03594.1 JAD03931.1 CH933809 EDW19398.1 GAKP01011308 GAKP01011306 GAKP01011305 JAC47647.1 GFDF01004042 JAV10042.1 GFDF01004043 JAV10041.1 GAMC01009758 GAMC01009757 JAB96797.1 GDHF01031724 GDHF01026089 GDHF01018165 GDHF01013860 GDHF01002052 JAI20590.1 JAI26225.1 JAI34149.1 JAI38454.1 JAI50262.1 GALA01000137 JAA94715.1 LBMM01002047 KMQ95354.1 GGFM01001479 MBW22230.1 KR058818 AKB96223.1 KR058819 AKB96224.1 KR058820 AKB96225.1 GGFK01002134 MBW35455.1 KA646958 AFP61587.1 GALA01001243 JAA93609.1 GDRN01062059 JAI65116.1 ATLV01013899 ATLV01013900 KE524908 KFB38249.1 CH963847 KRF97623.1 AAAB01008957 EAA10674.3 AHAT01001545 AERX01036628 AERX01036629 KY753873 AVC68725.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000235965
UP000027135
UP000007266
+ More
UP000030742 UP000019118 UP000078492 UP000078541 UP000008237 UP000007819 UP000008820 UP000009192 UP000192221 UP000036403 UP000095301 UP000076408 UP000030765 UP000095300 UP000075880 UP000007798 UP000007062 UP000018468 UP000261340 UP000261460 UP000261400 UP000264840 UP000261580 UP000265040 UP000265100 UP000265160 UP000261360 UP000261420 UP000264800 UP000007303 UP000261520 UP000261440 UP000261640 UP000005207 UP000257200
UP000030742 UP000019118 UP000078492 UP000078541 UP000008237 UP000007819 UP000008820 UP000009192 UP000192221 UP000036403 UP000095301 UP000076408 UP000030765 UP000095300 UP000075880 UP000007798 UP000007062 UP000018468 UP000261340 UP000261460 UP000261400 UP000264840 UP000261580 UP000265040 UP000265100 UP000265160 UP000261360 UP000261420 UP000264800 UP000007303 UP000261520 UP000261440 UP000261640 UP000005207 UP000257200
Pfam
PF00012 HSP70
Interpro
Gene 3D
ProteinModelPortal
H9IVD8
A0A2H1VDU5
A0A2A4IWH0
A0A2A4IX17
A0A194RH35
A0A2A4IVL4
+ More
S4PX99 A0A2J7QEY4 A0A1B6C8J9 A0A1B6FM09 A0A1B6GU52 A0A1Y1JTX9 A0A1Y1JQX1 A0A067R776 A0A346THM8 A0A224X7G5 D2A3E8 A0A023F9S1 V9IBC4 U4U4I6 N6UBE3 A0A195E535 A0A195F4P4 R4G605 E2B7Z4 A0A0A9XY29 A0A146L8M9 A0A0K8TST6 A0A0C9R4T8 A0A1B6FPX4 U5EM97 A0A1B6F6H7 A0A1I9WLB6 A0A336MFX0 A0A336M9B2 F2EJC0 J9JVZ3 A0A2S2QMS8 J9HSH6 A0A1Q3FWD8 A0A023EXV4 A0A2H8TT55 A0A2S2PFX9 A0A0A1WZ41 B4KV58 A0A034VZY9 A0A1L8DUC1 A0A1L8DUD5 W8BDM7 A0A1W4U6M6 A0A1W4UK43 A0A0K8U2L3 T1DGM2 A0A0J7KYH4 A0A2M3Z133 A0A0E3T149 A0A0E3T267 A0A1I8MN87 A0A0E3T0V0 A0A2M4A3X9 T1PGF6 A0A182YE12 T1D3U1 A0A0P4WCY3 A0A084VJV5 A0A1I8P985 A0A1I8P9G3 A0A182IL77 A0A0Q9WP46 Q7Q7J4 W5MXU8 A0A3Q0R6W2 A0A3B4ERF4 A0A3B5A539 A0A3Q2WA53 A0A3Q4H4C0 A0A3Q1HJZ1 A0A3P8NZ65 A0A3P9C0W6 A0A3B4XMS6 A0A3B4EPJ4 A0A3B4TQN4 A0A3Q2ZY40 A0A3Q2UTM5 H3CU62 A0A3B4AY19 A0A3B4XHI4 A0A3B4D9D0 A0A3Q3LS94 I3K531 A0A2L1DG12 A0A3Q1GC59 A0A3B5AES1
S4PX99 A0A2J7QEY4 A0A1B6C8J9 A0A1B6FM09 A0A1B6GU52 A0A1Y1JTX9 A0A1Y1JQX1 A0A067R776 A0A346THM8 A0A224X7G5 D2A3E8 A0A023F9S1 V9IBC4 U4U4I6 N6UBE3 A0A195E535 A0A195F4P4 R4G605 E2B7Z4 A0A0A9XY29 A0A146L8M9 A0A0K8TST6 A0A0C9R4T8 A0A1B6FPX4 U5EM97 A0A1B6F6H7 A0A1I9WLB6 A0A336MFX0 A0A336M9B2 F2EJC0 J9JVZ3 A0A2S2QMS8 J9HSH6 A0A1Q3FWD8 A0A023EXV4 A0A2H8TT55 A0A2S2PFX9 A0A0A1WZ41 B4KV58 A0A034VZY9 A0A1L8DUC1 A0A1L8DUD5 W8BDM7 A0A1W4U6M6 A0A1W4UK43 A0A0K8U2L3 T1DGM2 A0A0J7KYH4 A0A2M3Z133 A0A0E3T149 A0A0E3T267 A0A1I8MN87 A0A0E3T0V0 A0A2M4A3X9 T1PGF6 A0A182YE12 T1D3U1 A0A0P4WCY3 A0A084VJV5 A0A1I8P985 A0A1I8P9G3 A0A182IL77 A0A0Q9WP46 Q7Q7J4 W5MXU8 A0A3Q0R6W2 A0A3B4ERF4 A0A3B5A539 A0A3Q2WA53 A0A3Q4H4C0 A0A3Q1HJZ1 A0A3P8NZ65 A0A3P9C0W6 A0A3B4XMS6 A0A3B4EPJ4 A0A3B4TQN4 A0A3Q2ZY40 A0A3Q2UTM5 H3CU62 A0A3B4AY19 A0A3B4XHI4 A0A3B4D9D0 A0A3Q3LS94 I3K531 A0A2L1DG12 A0A3Q1GC59 A0A3B5AES1
PDB
6GFA
E-value=5.17073e-128,
Score=1175
Ontologies
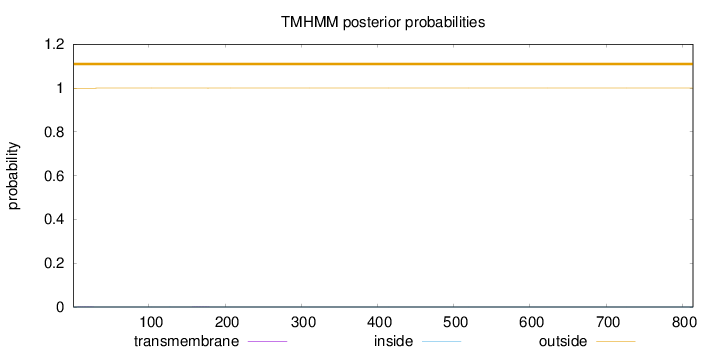
Topology
Length:
814
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01048
Exp number, first 60 AAs:
0.00409999999999999
Total prob of N-in:
0.00055
outside
1 - 814
Population Genetic Test Statistics
Pi
219.075721
Theta
180.981154
Tajima's D
0.597822
CLR
0
CSRT
0.540122993850307
Interpretation
Uncertain
