Gene
KWMTBOMO07819
Pre Gene Modal
BGIBMGA001219
Annotation
PREDICTED:_ethanolamine_kinase_[Papilio_machaon]
Full name
Ethanolamine kinase
Alternative Name
Protein easily shocked
Location in the cell
Cytoplasmic Reliability : 1.226 PlasmaMembrane Reliability : 1.108
Sequence
CDS
ATGCATAGAGTTGAACTTGGAAAAGAGGTACAAAAAGATCCAATGGTATGGGATAAAATTGAACAATTCCTAAGTTTGTTGCCAGAGAAATTCTCTACAGAAGTCAAACACAATAGATTTGTGAGTAGCTTTGGGTCAGTAACTAAGCTAAGGATTGAATTTGAGCGTCTAAAGTCTCATCTAATCAAAACGGAGAGCCCAATAGTGTTTGCTCACAATGATTTGCTTCTAGGAAACGTTATCTTCAACAAAGATGAAGGGACCATCTCTTTCATAGACTATGAATATGCCAGTTACAATTATCAGGCGTTCGATATAGCTAATCATTTCAACGAATTTGTCGGATTATCAATTGAAGACATTGATTACCAAAGATATCCGAGTAAAGACTTCCAACTTGCTTGGATTAGAGTATATTTATCGGAATATTTAGGAACAATGACGCCTGACGCTAAAAGCGTAGATACAGTTTATGAAGAAGTACAAAAACTTTCGTTAGCTTCACATTTTCTTTGGGGTATATGGTCACTAGTGCAATTTGAACATTCTGATATTGATTTCGATTTCGGAAGGTATGCAGAAATACGTTTACAGCGATATTATCAAACAAAAAATGATATTTTAAGTGTATGTACTTAA
Protein
MHRVELGKEVQKDPMVWDKIEQFLSLLPEKFSTEVKHNRFVSSFGSVTKLRIEFERLKSHLIKTESPIVFAHNDLLLGNVIFNKDEGTISFIDYEYASYNYQAFDIANHFNEFVGLSIEDIDYQRYPSKDFQLAWIRVYLSEYLGTMTPDAKSVDTVYEEVQKLSLASHFLWGIWSLVQFEHSDIDFDFGRYAEIRLQRYYQTKNDILSVCT
Summary
Description
Highly specific for ethanolamine phosphorylation. May be a rate-controlling step in phosphatidylethanolamine biosynthesis.
Catalytic Activity
ATP + ethanolamine = ADP + H(+) + phosphoethanolamine
Similarity
Belongs to the choline/ethanolamine kinase family.
Keywords
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
Kinase
Lipid biosynthesis
Lipid metabolism
Nucleotide-binding
Phospholipid biosynthesis
Phospholipid metabolism
Phosphoprotein
Reference proteome
Transferase
Feature
chain Ethanolamine kinase
splice variant In isoform G.
splice variant In isoform G.
Uniprot
H9IVD9
A0A194RHT6
A0A2H1VLK0
A0A2A4J114
A0A194PE10
A0A0L7LJC1
+ More
A0A212EME1 A0A1B0DLT2 A0A067QIM2 A0A1L8E3P9 A0A0K8TLL7 Q16UX5 A0A1Y1N783 A0A182P7L7 A0A182W4T5 A0A2J7PXA4 A0A1L8ED31 A0A182M9N0 A0A182UMP1 A0A1Q3FJ61 Q29GE0 A0A0A1X0W6 W8BJB8 B0WH47 A0A182T605 A0A182KY26 Q7QEI8 A0A1A9VWK0 A0A1B0A1D6 A0A1B0BLH5 A0A1A9YQC7 A0A3B0KE78 A0A182RHV2 A0A1B6MNL2 V5GUQ1 A0A336MXR1 B3N0U6 A0A1B6IDX1 A0A182UB63 A0A182YK58 T1PFM6 A0A0L0BW36 B4NEW5 A0A0M5J366 A0A1B6DMU5 A0A182X582 A0A1B0FCR2 A0A2M4BT60 B4L7Q0 A0A2M4BRA6 A0A2M4BR65 T1GU53 B4JJY2 P54352-2 D6X535 P54352 A0A182N908 A0A0K8TZU6 A0A023ETY5 B4PXQ8 B3NXD9 A0A023EMT2 B4MG95 A0A0R1EAE6 A0A182G1P3 A0A0Q5T4K5 A0A1W4WDE5 A0A1I8PK72 A0A1B6CIE0 A0A0K8WCP8 A0A2M3ZAX1 A0A1A9WHQ5 A0A182FM71 A0A151WRR4 A0A158NE75 A0A182Q509 A0A1B0C9X0 A0A182I9Q4 A0A0N0BCT5 A0A088A9M7 A0A151IDI5 A0A034VAH8 A0A182IME6 B4NUI5 A0A034V885 A0A154PDA3 A0A0L7RFN0 W5JVX3 F4X444 A0A1W4XEF8 E9IA48 A0A3R7PLU0 A0A1S6KZC0 A0A3P8ZZ15 A0A232FBQ7 E0W045 A0A1W5AEP7 A0A0T6B455 M3XLG4
A0A212EME1 A0A1B0DLT2 A0A067QIM2 A0A1L8E3P9 A0A0K8TLL7 Q16UX5 A0A1Y1N783 A0A182P7L7 A0A182W4T5 A0A2J7PXA4 A0A1L8ED31 A0A182M9N0 A0A182UMP1 A0A1Q3FJ61 Q29GE0 A0A0A1X0W6 W8BJB8 B0WH47 A0A182T605 A0A182KY26 Q7QEI8 A0A1A9VWK0 A0A1B0A1D6 A0A1B0BLH5 A0A1A9YQC7 A0A3B0KE78 A0A182RHV2 A0A1B6MNL2 V5GUQ1 A0A336MXR1 B3N0U6 A0A1B6IDX1 A0A182UB63 A0A182YK58 T1PFM6 A0A0L0BW36 B4NEW5 A0A0M5J366 A0A1B6DMU5 A0A182X582 A0A1B0FCR2 A0A2M4BT60 B4L7Q0 A0A2M4BRA6 A0A2M4BR65 T1GU53 B4JJY2 P54352-2 D6X535 P54352 A0A182N908 A0A0K8TZU6 A0A023ETY5 B4PXQ8 B3NXD9 A0A023EMT2 B4MG95 A0A0R1EAE6 A0A182G1P3 A0A0Q5T4K5 A0A1W4WDE5 A0A1I8PK72 A0A1B6CIE0 A0A0K8WCP8 A0A2M3ZAX1 A0A1A9WHQ5 A0A182FM71 A0A151WRR4 A0A158NE75 A0A182Q509 A0A1B0C9X0 A0A182I9Q4 A0A0N0BCT5 A0A088A9M7 A0A151IDI5 A0A034VAH8 A0A182IME6 B4NUI5 A0A034V885 A0A154PDA3 A0A0L7RFN0 W5JVX3 F4X444 A0A1W4XEF8 E9IA48 A0A3R7PLU0 A0A1S6KZC0 A0A3P8ZZ15 A0A232FBQ7 E0W045 A0A1W5AEP7 A0A0T6B455 M3XLG4
EC Number
2.7.1.82
Pubmed
19121390
26354079
26227816
22118469
24845553
26369729
+ More
17510324 28004739 15632085 17994087 23185243 25830018 24495485 20966253 12364791 14747013 17210077 18057021 25244985 25315136 26108605 7923374 10731132 12537572 12537569 17372656 18327897 18362917 19820115 24945155 17550304 26483478 21347285 25348373 20920257 23761445 21719571 21282665 24330026 25069045 28648823 20566863 9215903
17510324 28004739 15632085 17994087 23185243 25830018 24495485 20966253 12364791 14747013 17210077 18057021 25244985 25315136 26108605 7923374 10731132 12537572 12537569 17372656 18327897 18362917 19820115 24945155 17550304 26483478 21347285 25348373 20920257 23761445 21719571 21282665 24330026 25069045 28648823 20566863 9215903
EMBL
BABH01000950
BABH01000951
BABH01000952
BABH01000953
KQ460205
KPJ16890.1
+ More
ODYU01003229 SOQ41710.1 NWSH01004427 PCG65130.1 KQ459606 KPI91263.1 JTDY01000893 KOB75530.1 AGBW02013857 OWR42647.1 AJVK01006808 KK853502 KDR07130.1 GFDF01000820 JAV13264.1 GDAI01002545 JAI15058.1 CH477612 EAT38330.1 GEZM01010845 GEZM01010844 GEZM01010843 JAV93772.1 NEVH01020859 PNF20957.1 GFDG01002149 JAV16650.1 AXCM01006140 GFDL01007522 JAV27523.1 CH379064 EAL32170.3 KRT07016.1 KRT07017.1 GBXI01009308 JAD04984.1 GAMC01007720 GAMC01007718 JAB98837.1 DS231932 EDS27498.1 AAAB01008846 EAA06461.5 JXJN01016356 OUUW01000011 SPP86610.1 GEBQ01002484 JAT37493.1 GALX01004528 JAB63938.1 UFQS01003664 UFQT01003664 SSX15847.1 SSX35192.1 CH902644 EDV34825.1 KPU75301.1 KPU75302.1 GECU01022580 GECU01018182 JAS85126.1 JAS89524.1 KA646708 AFP61337.1 JRES01001250 KNC24231.1 CH964239 EDW82284.2 CP012528 ALC49248.1 GEDC01015125 GEDC01010287 JAS22173.1 JAS27011.1 CCAG010001428 GGFJ01007124 MBW56265.1 CH933814 EDW05763.1 KRG07515.1 KRG07516.1 GGFJ01006372 MBW55513.1 GGFJ01006371 MBW55512.1 CAQQ02139792 CAQQ02139793 CH916370 EDV99884.1 L35603 L35604 AE014298 AY118437 BT025059 BT099812 KQ971382 EEZ97183.2 GDHF01032709 JAI19605.1 GAPW01001704 JAC11894.1 CM000162 EDX01894.2 KRK06419.1 KRK06420.1 KRK06421.1 CH954180 EDV46898.2 KQS30237.1 GAPW01002925 JAC10673.1 CH940672 EDW57418.1 KRF77765.1 KRK06422.1 JXUM01137480 KQ568584 KXJ68949.1 KQS30236.1 GEDC01024111 JAS13187.1 GDHF01016024 GDHF01003356 JAI36290.1 JAI48958.1 GGFM01004901 MBW25652.1 KQ982796 KYQ50602.1 ADTU01013096 ADTU01013097 AXCN02001403 AJWK01002980 APCN01001018 KQ435898 KOX69172.1 KQ977939 KYM98526.1 GAKP01019428 JAC39524.1 AXCP01008829 CH984024 EDX16632.1 GAKP01019426 JAC39526.1 KQ434869 KZC09238.1 KQ414606 KOC69613.1 ADMH02000145 ETN67618.1 GL888625 EGI58768.1 GL761987 EFZ22558.1 QCYY01001692 ROT76128.1 KC223178 AQT27081.1 NNAY01000455 OXU28274.1 DS235855 EEB19001.1 LJIG01009900 KRT82156.1 AFYH01035462 AFYH01035463 AFYH01035464 AFYH01035465 AFYH01035466 AFYH01035467 AFYH01035468 AFYH01035469 AFYH01035470 AFYH01035471
ODYU01003229 SOQ41710.1 NWSH01004427 PCG65130.1 KQ459606 KPI91263.1 JTDY01000893 KOB75530.1 AGBW02013857 OWR42647.1 AJVK01006808 KK853502 KDR07130.1 GFDF01000820 JAV13264.1 GDAI01002545 JAI15058.1 CH477612 EAT38330.1 GEZM01010845 GEZM01010844 GEZM01010843 JAV93772.1 NEVH01020859 PNF20957.1 GFDG01002149 JAV16650.1 AXCM01006140 GFDL01007522 JAV27523.1 CH379064 EAL32170.3 KRT07016.1 KRT07017.1 GBXI01009308 JAD04984.1 GAMC01007720 GAMC01007718 JAB98837.1 DS231932 EDS27498.1 AAAB01008846 EAA06461.5 JXJN01016356 OUUW01000011 SPP86610.1 GEBQ01002484 JAT37493.1 GALX01004528 JAB63938.1 UFQS01003664 UFQT01003664 SSX15847.1 SSX35192.1 CH902644 EDV34825.1 KPU75301.1 KPU75302.1 GECU01022580 GECU01018182 JAS85126.1 JAS89524.1 KA646708 AFP61337.1 JRES01001250 KNC24231.1 CH964239 EDW82284.2 CP012528 ALC49248.1 GEDC01015125 GEDC01010287 JAS22173.1 JAS27011.1 CCAG010001428 GGFJ01007124 MBW56265.1 CH933814 EDW05763.1 KRG07515.1 KRG07516.1 GGFJ01006372 MBW55513.1 GGFJ01006371 MBW55512.1 CAQQ02139792 CAQQ02139793 CH916370 EDV99884.1 L35603 L35604 AE014298 AY118437 BT025059 BT099812 KQ971382 EEZ97183.2 GDHF01032709 JAI19605.1 GAPW01001704 JAC11894.1 CM000162 EDX01894.2 KRK06419.1 KRK06420.1 KRK06421.1 CH954180 EDV46898.2 KQS30237.1 GAPW01002925 JAC10673.1 CH940672 EDW57418.1 KRF77765.1 KRK06422.1 JXUM01137480 KQ568584 KXJ68949.1 KQS30236.1 GEDC01024111 JAS13187.1 GDHF01016024 GDHF01003356 JAI36290.1 JAI48958.1 GGFM01004901 MBW25652.1 KQ982796 KYQ50602.1 ADTU01013096 ADTU01013097 AXCN02001403 AJWK01002980 APCN01001018 KQ435898 KOX69172.1 KQ977939 KYM98526.1 GAKP01019428 JAC39524.1 AXCP01008829 CH984024 EDX16632.1 GAKP01019426 JAC39526.1 KQ434869 KZC09238.1 KQ414606 KOC69613.1 ADMH02000145 ETN67618.1 GL888625 EGI58768.1 GL761987 EFZ22558.1 QCYY01001692 ROT76128.1 KC223178 AQT27081.1 NNAY01000455 OXU28274.1 DS235855 EEB19001.1 LJIG01009900 KRT82156.1 AFYH01035462 AFYH01035463 AFYH01035464 AFYH01035465 AFYH01035466 AFYH01035467 AFYH01035468 AFYH01035469 AFYH01035470 AFYH01035471
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000037510
UP000007151
+ More
UP000092462 UP000027135 UP000008820 UP000075885 UP000075920 UP000235965 UP000075883 UP000075903 UP000001819 UP000002320 UP000075901 UP000075882 UP000007062 UP000078200 UP000092445 UP000092460 UP000092443 UP000268350 UP000075900 UP000007801 UP000075902 UP000076408 UP000095301 UP000037069 UP000007798 UP000092553 UP000076407 UP000092444 UP000009192 UP000015102 UP000001070 UP000000803 UP000007266 UP000075884 UP000002282 UP000008711 UP000008792 UP000069940 UP000249989 UP000192221 UP000095300 UP000091820 UP000069272 UP000075809 UP000005205 UP000075886 UP000092461 UP000075840 UP000053105 UP000005203 UP000078542 UP000075880 UP000000304 UP000076502 UP000053825 UP000000673 UP000007755 UP000192223 UP000283509 UP000265140 UP000215335 UP000009046 UP000192224 UP000008672
UP000092462 UP000027135 UP000008820 UP000075885 UP000075920 UP000235965 UP000075883 UP000075903 UP000001819 UP000002320 UP000075901 UP000075882 UP000007062 UP000078200 UP000092445 UP000092460 UP000092443 UP000268350 UP000075900 UP000007801 UP000075902 UP000076408 UP000095301 UP000037069 UP000007798 UP000092553 UP000076407 UP000092444 UP000009192 UP000015102 UP000001070 UP000000803 UP000007266 UP000075884 UP000002282 UP000008711 UP000008792 UP000069940 UP000249989 UP000192221 UP000095300 UP000091820 UP000069272 UP000075809 UP000005205 UP000075886 UP000092461 UP000075840 UP000053105 UP000005203 UP000078542 UP000075880 UP000000304 UP000076502 UP000053825 UP000000673 UP000007755 UP000192223 UP000283509 UP000265140 UP000215335 UP000009046 UP000192224 UP000008672
Interpro
CDD
ProteinModelPortal
H9IVD9
A0A194RHT6
A0A2H1VLK0
A0A2A4J114
A0A194PE10
A0A0L7LJC1
+ More
A0A212EME1 A0A1B0DLT2 A0A067QIM2 A0A1L8E3P9 A0A0K8TLL7 Q16UX5 A0A1Y1N783 A0A182P7L7 A0A182W4T5 A0A2J7PXA4 A0A1L8ED31 A0A182M9N0 A0A182UMP1 A0A1Q3FJ61 Q29GE0 A0A0A1X0W6 W8BJB8 B0WH47 A0A182T605 A0A182KY26 Q7QEI8 A0A1A9VWK0 A0A1B0A1D6 A0A1B0BLH5 A0A1A9YQC7 A0A3B0KE78 A0A182RHV2 A0A1B6MNL2 V5GUQ1 A0A336MXR1 B3N0U6 A0A1B6IDX1 A0A182UB63 A0A182YK58 T1PFM6 A0A0L0BW36 B4NEW5 A0A0M5J366 A0A1B6DMU5 A0A182X582 A0A1B0FCR2 A0A2M4BT60 B4L7Q0 A0A2M4BRA6 A0A2M4BR65 T1GU53 B4JJY2 P54352-2 D6X535 P54352 A0A182N908 A0A0K8TZU6 A0A023ETY5 B4PXQ8 B3NXD9 A0A023EMT2 B4MG95 A0A0R1EAE6 A0A182G1P3 A0A0Q5T4K5 A0A1W4WDE5 A0A1I8PK72 A0A1B6CIE0 A0A0K8WCP8 A0A2M3ZAX1 A0A1A9WHQ5 A0A182FM71 A0A151WRR4 A0A158NE75 A0A182Q509 A0A1B0C9X0 A0A182I9Q4 A0A0N0BCT5 A0A088A9M7 A0A151IDI5 A0A034VAH8 A0A182IME6 B4NUI5 A0A034V885 A0A154PDA3 A0A0L7RFN0 W5JVX3 F4X444 A0A1W4XEF8 E9IA48 A0A3R7PLU0 A0A1S6KZC0 A0A3P8ZZ15 A0A232FBQ7 E0W045 A0A1W5AEP7 A0A0T6B455 M3XLG4
A0A212EME1 A0A1B0DLT2 A0A067QIM2 A0A1L8E3P9 A0A0K8TLL7 Q16UX5 A0A1Y1N783 A0A182P7L7 A0A182W4T5 A0A2J7PXA4 A0A1L8ED31 A0A182M9N0 A0A182UMP1 A0A1Q3FJ61 Q29GE0 A0A0A1X0W6 W8BJB8 B0WH47 A0A182T605 A0A182KY26 Q7QEI8 A0A1A9VWK0 A0A1B0A1D6 A0A1B0BLH5 A0A1A9YQC7 A0A3B0KE78 A0A182RHV2 A0A1B6MNL2 V5GUQ1 A0A336MXR1 B3N0U6 A0A1B6IDX1 A0A182UB63 A0A182YK58 T1PFM6 A0A0L0BW36 B4NEW5 A0A0M5J366 A0A1B6DMU5 A0A182X582 A0A1B0FCR2 A0A2M4BT60 B4L7Q0 A0A2M4BRA6 A0A2M4BR65 T1GU53 B4JJY2 P54352-2 D6X535 P54352 A0A182N908 A0A0K8TZU6 A0A023ETY5 B4PXQ8 B3NXD9 A0A023EMT2 B4MG95 A0A0R1EAE6 A0A182G1P3 A0A0Q5T4K5 A0A1W4WDE5 A0A1I8PK72 A0A1B6CIE0 A0A0K8WCP8 A0A2M3ZAX1 A0A1A9WHQ5 A0A182FM71 A0A151WRR4 A0A158NE75 A0A182Q509 A0A1B0C9X0 A0A182I9Q4 A0A0N0BCT5 A0A088A9M7 A0A151IDI5 A0A034VAH8 A0A182IME6 B4NUI5 A0A034V885 A0A154PDA3 A0A0L7RFN0 W5JVX3 F4X444 A0A1W4XEF8 E9IA48 A0A3R7PLU0 A0A1S6KZC0 A0A3P8ZZ15 A0A232FBQ7 E0W045 A0A1W5AEP7 A0A0T6B455 M3XLG4
PDB
4DA5
E-value=1.55616e-26,
Score=293
Ontologies
PATHWAY
GO
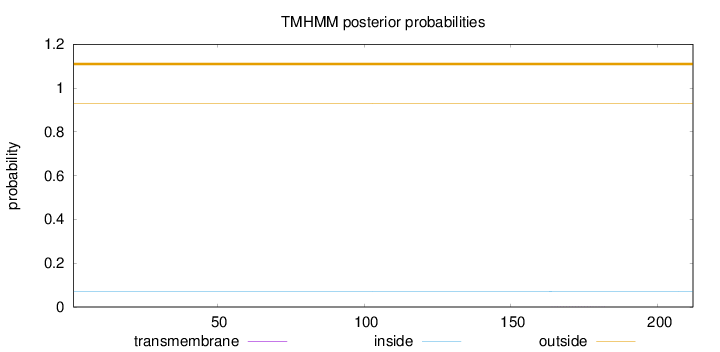
Topology
Subcellular location
Cytoplasm
Length:
212
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00736
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.07174
outside
1 - 212
Population Genetic Test Statistics
Pi
224.282757
Theta
164.577886
Tajima's D
0.494004
CLR
1.78919
CSRT
0.511974401279936
Interpretation
Uncertain
