Gene
KWMTBOMO07814 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000923
Annotation
PREDICTED:_cytochrome_b-c1_complex_subunit_8_[Plutella_xylostella]
Location in the cell
Mitochondrial Reliability : 2.271
Sequence
CDS
ATGGGCAAACACTTTGGAGAACTAGCTAAAATTCGGGGTCTTGTCACCTACAAGATATCTGCCCATGAACAACGAGCATATGCTGGAGCTATTTCAAATGGTATTCCTAATATCTTCAGAAGATTTCGTGAGAGCGTTTTTAAAGTAGTCCCGCCCTTCATCATTGGATACTTGATATATGAAGGAGTTGAAAGGGAGCATCACAGACTATCACGGAAGAACCCTGCTGATTTTGAAAATGATCAATAA
Protein
MGKHFGELAKIRGLVTYKISAHEQRAYAGAISNGIPNIFRRFRESVFKVVPPFIIGYLIYEGVEREHHRLSRKNPADFENDQ
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
H9IUJ3
A0A2H1VN79
A0A3S2LBK1
A0A0L7K3R0
A0A212EME6
I4DJF7
+ More
A0A194RHH4 S4NXC4 A0A1B0DCH8 A0A1B0FBL5 A0A1A9UJ68 A0A1I8QF14 A0A1I8MSA3 A0A3B0K1X4 A0A0M4ECF8 B4N6F4 A0A1B0A6K2 A0A1L8EF75 A0A1L8E0W6 Q2LZI3 A0A2J7Q503 B4H6N0 A0A0L0BV84 A0A1W4W4I6 A0A1L8EHX6 U5EWU3 B4IX18 B3M4I9 B4HKT0 B4QNV5 Q9VVH5 B4PJU9 B3NHU8 B4LFR8 B4KXE7 Q1W286 A0A1B6MB62 A0A1J1HWE0 A0A336LYN2 D1FQ43 A0A1W4X507 B5M0W3 A0A2M3Z047 A0A2M4A6B9 W5JLZ0 A0A2C9JTQ1 A0A224ZB31 A0A131Z597 A0A293LYV3 A0A093IPX6 A0A1L8E9I8 A0A2M4A802 D2A1S3 A0A087UF51 A0A2P6L8V1 C9W1P3 J9HFH0 A0A182G7Z1 Q7QDP2 A0A182R929 A0A3F2Z216 A0A1S4H4M9 A0A0P5TBY9 A0A084VJW1 A0A182IKK3 A0A182QGQ1 A0A094KVC3 A0A091KRL1 Q6Q9G3 K4G6V8 A0A023GEA3 A0A226E260 T1DJW3 A0A2I0T386 A0A091LNX2 A0A1B0AY58 B0WZV5 A0A182FZS5 Q09JK0 A0A0C9RZW0 Q4PM49 A0A147BVR0 B7PRC5 A0A1D2MD05 A0A3S1AQU9 A0A091FLJ2 A0A0P4YPP2 A0A091Q0X4 A0A1V4KTY2 A0A023FG99 A0A023FES7 W4ZFW5 A0A182HIW0 A0A1Q3FLY5 A0A3Q3LFP9 A0A091T0U1 A0A091S8P1 E9FT85 A0A0B7B1F4 A0A2I0LT61
A0A194RHH4 S4NXC4 A0A1B0DCH8 A0A1B0FBL5 A0A1A9UJ68 A0A1I8QF14 A0A1I8MSA3 A0A3B0K1X4 A0A0M4ECF8 B4N6F4 A0A1B0A6K2 A0A1L8EF75 A0A1L8E0W6 Q2LZI3 A0A2J7Q503 B4H6N0 A0A0L0BV84 A0A1W4W4I6 A0A1L8EHX6 U5EWU3 B4IX18 B3M4I9 B4HKT0 B4QNV5 Q9VVH5 B4PJU9 B3NHU8 B4LFR8 B4KXE7 Q1W286 A0A1B6MB62 A0A1J1HWE0 A0A336LYN2 D1FQ43 A0A1W4X507 B5M0W3 A0A2M3Z047 A0A2M4A6B9 W5JLZ0 A0A2C9JTQ1 A0A224ZB31 A0A131Z597 A0A293LYV3 A0A093IPX6 A0A1L8E9I8 A0A2M4A802 D2A1S3 A0A087UF51 A0A2P6L8V1 C9W1P3 J9HFH0 A0A182G7Z1 Q7QDP2 A0A182R929 A0A3F2Z216 A0A1S4H4M9 A0A0P5TBY9 A0A084VJW1 A0A182IKK3 A0A182QGQ1 A0A094KVC3 A0A091KRL1 Q6Q9G3 K4G6V8 A0A023GEA3 A0A226E260 T1DJW3 A0A2I0T386 A0A091LNX2 A0A1B0AY58 B0WZV5 A0A182FZS5 Q09JK0 A0A0C9RZW0 Q4PM49 A0A147BVR0 B7PRC5 A0A1D2MD05 A0A3S1AQU9 A0A091FLJ2 A0A0P4YPP2 A0A091Q0X4 A0A1V4KTY2 A0A023FG99 A0A023FES7 W4ZFW5 A0A182HIW0 A0A1Q3FLY5 A0A3Q3LFP9 A0A091T0U1 A0A091S8P1 E9FT85 A0A0B7B1F4 A0A2I0LT61
Pubmed
19121390
26227816
22118469
22651552
26354079
23622113
+ More
25315136 17994087 18057021 15632085 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 19166301 20920257 23761445 15562597 28797301 26830274 18362917 19820115 20650005 24029695 17510324 26483478 12364791 24438588 23056606 24330624 18070664 26131772 16431279 29652888 27289101 21292972 23371554
25315136 17994087 18057021 15632085 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 19166301 20920257 23761445 15562597 28797301 26830274 18362917 19820115 20650005 24029695 17510324 26483478 12364791 24438588 23056606 24330624 18070664 26131772 16431279 29652888 27289101 21292972 23371554
EMBL
BABH01000961
BABH01000962
ODYU01003229
SOQ41714.1
RSAL01000276
RVE43109.1
+ More
JTDY01011558 KOB55595.1 AGBW02013857 OWR42646.1 AK401425 KQ459606 BAM18047.1 KPI91262.1 KQ460205 KPJ16889.1 GAIX01009109 JAA83451.1 AJVK01005095 CCAG010008960 OUUW01000012 SPP87323.1 CP012525 ALC43096.1 CH964154 EDW79943.1 KRF99194.1 KRF99195.1 GFDG01001475 JAV17324.1 GFDF01001780 JAV12304.1 CH379069 EAL29524.1 NEVH01018372 PNF23663.1 CH479214 EDW33458.1 JRES01001279 KNC23975.1 GFDG01000573 JAV18226.1 GANO01001264 JAB58607.1 CH916366 EDV96324.1 CH902618 EDV40483.1 KPU78585.1 CH480815 EDW41885.1 CM000363 CM002912 EDX10857.1 KMZ00237.1 AE014296 AY113252 KX531481 AAF49336.1 AAM29257.1 ADV37560.1 ADV37561.1 AGB94680.1 ANY27291.1 CM000159 EDW94718.1 KRK02013.1 CH954178 EDV51963.1 CH940647 EDW69295.1 CH933809 EDW18633.1 DQ445541 ABD98779.1 GEBQ01006807 JAT33170.1 CVRI01000021 CRK91676.1 UFQT01000309 SSX23092.1 EZ419943 ACZ28298.1 EU930299 ACH56927.1 GGFM01001151 MBW21902.1 GGFK01002941 MBW36262.1 ADMH02000671 ETN65382.1 GFPF01013038 MAA24184.1 GEDV01002622 JAP85935.1 GFWV01008935 MAA33664.1 KL216278 KFV68746.1 GFDG01003401 JAV15398.1 GGFK01003541 MBW36862.1 KQ971338 EFA02862.1 KK119549 KFM75990.1 MWRG01000923 PRD34985.1 EZ406195 ACX53990.1 CH477328 EJY57567.1 JXUM01047141 KQ561507 KXJ78355.1 AAAB01008849 EAA07167.2 GDIP01204056 GDIP01134264 GDIQ01072326 JAL69450.1 JAN22411.1 ATLV01013905 KE524908 KFB38255.1 AXCN02002267 KL356584 KFZ62570.1 KK539336 KFP30409.1 AY552057 AAS79343.1 JX052744 JX209397 AFK10972.1 AFM87711.1 GBBM01003216 GBBM01003215 JAC32202.1 LNIX01000007 OXA51835.1 GALA01000021 JAA94831.1 KZ521456 PKU28270.1 KK504388 KFP60836.1 JXJN01005553 JXJN01005554 JXJN01005555 DS232218 EDS37780.1 DQ886846 ABI52763.1 GBZX01002931 JAG89809.1 DQ066278 AAY66915.1 GEGO01000561 JAR94843.1 ABJB010815611 DS771975 EEC09147.1 LJIJ01001766 ODM90855.1 RQTK01001663 RUS69499.1 KL447125 KFO70034.1 GDIP01224605 JAI98796.1 KK653539 KFQ03319.1 LSYS01001584 OPJ87904.1 GBBK01004478 JAC20004.1 GBBK01004480 JAC20002.1 AAGJ04042127 APCN01006664 APCN01006665 GFDL01006549 JAV28496.1 KK939647 KFQ49714.1 KK812809 KFQ36810.1 GL732524 EFX89330.1 HACG01039271 CEK86136.1 AKCR02000104 PKK20618.1
JTDY01011558 KOB55595.1 AGBW02013857 OWR42646.1 AK401425 KQ459606 BAM18047.1 KPI91262.1 KQ460205 KPJ16889.1 GAIX01009109 JAA83451.1 AJVK01005095 CCAG010008960 OUUW01000012 SPP87323.1 CP012525 ALC43096.1 CH964154 EDW79943.1 KRF99194.1 KRF99195.1 GFDG01001475 JAV17324.1 GFDF01001780 JAV12304.1 CH379069 EAL29524.1 NEVH01018372 PNF23663.1 CH479214 EDW33458.1 JRES01001279 KNC23975.1 GFDG01000573 JAV18226.1 GANO01001264 JAB58607.1 CH916366 EDV96324.1 CH902618 EDV40483.1 KPU78585.1 CH480815 EDW41885.1 CM000363 CM002912 EDX10857.1 KMZ00237.1 AE014296 AY113252 KX531481 AAF49336.1 AAM29257.1 ADV37560.1 ADV37561.1 AGB94680.1 ANY27291.1 CM000159 EDW94718.1 KRK02013.1 CH954178 EDV51963.1 CH940647 EDW69295.1 CH933809 EDW18633.1 DQ445541 ABD98779.1 GEBQ01006807 JAT33170.1 CVRI01000021 CRK91676.1 UFQT01000309 SSX23092.1 EZ419943 ACZ28298.1 EU930299 ACH56927.1 GGFM01001151 MBW21902.1 GGFK01002941 MBW36262.1 ADMH02000671 ETN65382.1 GFPF01013038 MAA24184.1 GEDV01002622 JAP85935.1 GFWV01008935 MAA33664.1 KL216278 KFV68746.1 GFDG01003401 JAV15398.1 GGFK01003541 MBW36862.1 KQ971338 EFA02862.1 KK119549 KFM75990.1 MWRG01000923 PRD34985.1 EZ406195 ACX53990.1 CH477328 EJY57567.1 JXUM01047141 KQ561507 KXJ78355.1 AAAB01008849 EAA07167.2 GDIP01204056 GDIP01134264 GDIQ01072326 JAL69450.1 JAN22411.1 ATLV01013905 KE524908 KFB38255.1 AXCN02002267 KL356584 KFZ62570.1 KK539336 KFP30409.1 AY552057 AAS79343.1 JX052744 JX209397 AFK10972.1 AFM87711.1 GBBM01003216 GBBM01003215 JAC32202.1 LNIX01000007 OXA51835.1 GALA01000021 JAA94831.1 KZ521456 PKU28270.1 KK504388 KFP60836.1 JXJN01005553 JXJN01005554 JXJN01005555 DS232218 EDS37780.1 DQ886846 ABI52763.1 GBZX01002931 JAG89809.1 DQ066278 AAY66915.1 GEGO01000561 JAR94843.1 ABJB010815611 DS771975 EEC09147.1 LJIJ01001766 ODM90855.1 RQTK01001663 RUS69499.1 KL447125 KFO70034.1 GDIP01224605 JAI98796.1 KK653539 KFQ03319.1 LSYS01001584 OPJ87904.1 GBBK01004478 JAC20004.1 GBBK01004480 JAC20002.1 AAGJ04042127 APCN01006664 APCN01006665 GFDL01006549 JAV28496.1 KK939647 KFQ49714.1 KK812809 KFQ36810.1 GL732524 EFX89330.1 HACG01039271 CEK86136.1 AKCR02000104 PKK20618.1
Proteomes
UP000005204
UP000283053
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000092462 UP000092444 UP000078200 UP000095300 UP000095301 UP000268350 UP000092553 UP000007798 UP000092445 UP000001819 UP000235965 UP000008744 UP000037069 UP000192221 UP000001070 UP000007801 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000008792 UP000009192 UP000183832 UP000192223 UP000000673 UP000076420 UP000053875 UP000007266 UP000054359 UP000008820 UP000069940 UP000249989 UP000007062 UP000075900 UP000076407 UP000030765 UP000075880 UP000075886 UP000198287 UP000092460 UP000002320 UP000069272 UP000001555 UP000094527 UP000271974 UP000053760 UP000190648 UP000007110 UP000075840 UP000261640 UP000000305 UP000053872
UP000092462 UP000092444 UP000078200 UP000095300 UP000095301 UP000268350 UP000092553 UP000007798 UP000092445 UP000001819 UP000235965 UP000008744 UP000037069 UP000192221 UP000001070 UP000007801 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000008792 UP000009192 UP000183832 UP000192223 UP000000673 UP000076420 UP000053875 UP000007266 UP000054359 UP000008820 UP000069940 UP000249989 UP000007062 UP000075900 UP000076407 UP000030765 UP000075880 UP000075886 UP000198287 UP000092460 UP000002320 UP000069272 UP000001555 UP000094527 UP000271974 UP000053760 UP000190648 UP000007110 UP000075840 UP000261640 UP000000305 UP000053872
Interpro
IPR036642
Cyt_bc1_su8_sf
+ More
IPR004205 Cyt_bc1_su8
IPR006201 Neur_channel
IPR036719 Neuro-gated_channel_TM_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006028 GABAA/Glycine_rcpt
IPR036734 Neur_chan_lig-bd_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006202 Neur_chan_lig-bd
IPR013815 ATP_grasp_subdomain_1
IPR004344 TTL/TTLL_fam
IPR027750 TTLL1
IPR004205 Cyt_bc1_su8
IPR006201 Neur_channel
IPR036719 Neuro-gated_channel_TM_sf
IPR006029 Neurotrans-gated_channel_TM
IPR006028 GABAA/Glycine_rcpt
IPR036734 Neur_chan_lig-bd_sf
IPR018000 Neurotransmitter_ion_chnl_CS
IPR006202 Neur_chan_lig-bd
IPR013815 ATP_grasp_subdomain_1
IPR004344 TTL/TTLL_fam
IPR027750 TTLL1
Gene 3D
ProteinModelPortal
H9IUJ3
A0A2H1VN79
A0A3S2LBK1
A0A0L7K3R0
A0A212EME6
I4DJF7
+ More
A0A194RHH4 S4NXC4 A0A1B0DCH8 A0A1B0FBL5 A0A1A9UJ68 A0A1I8QF14 A0A1I8MSA3 A0A3B0K1X4 A0A0M4ECF8 B4N6F4 A0A1B0A6K2 A0A1L8EF75 A0A1L8E0W6 Q2LZI3 A0A2J7Q503 B4H6N0 A0A0L0BV84 A0A1W4W4I6 A0A1L8EHX6 U5EWU3 B4IX18 B3M4I9 B4HKT0 B4QNV5 Q9VVH5 B4PJU9 B3NHU8 B4LFR8 B4KXE7 Q1W286 A0A1B6MB62 A0A1J1HWE0 A0A336LYN2 D1FQ43 A0A1W4X507 B5M0W3 A0A2M3Z047 A0A2M4A6B9 W5JLZ0 A0A2C9JTQ1 A0A224ZB31 A0A131Z597 A0A293LYV3 A0A093IPX6 A0A1L8E9I8 A0A2M4A802 D2A1S3 A0A087UF51 A0A2P6L8V1 C9W1P3 J9HFH0 A0A182G7Z1 Q7QDP2 A0A182R929 A0A3F2Z216 A0A1S4H4M9 A0A0P5TBY9 A0A084VJW1 A0A182IKK3 A0A182QGQ1 A0A094KVC3 A0A091KRL1 Q6Q9G3 K4G6V8 A0A023GEA3 A0A226E260 T1DJW3 A0A2I0T386 A0A091LNX2 A0A1B0AY58 B0WZV5 A0A182FZS5 Q09JK0 A0A0C9RZW0 Q4PM49 A0A147BVR0 B7PRC5 A0A1D2MD05 A0A3S1AQU9 A0A091FLJ2 A0A0P4YPP2 A0A091Q0X4 A0A1V4KTY2 A0A023FG99 A0A023FES7 W4ZFW5 A0A182HIW0 A0A1Q3FLY5 A0A3Q3LFP9 A0A091T0U1 A0A091S8P1 E9FT85 A0A0B7B1F4 A0A2I0LT61
A0A194RHH4 S4NXC4 A0A1B0DCH8 A0A1B0FBL5 A0A1A9UJ68 A0A1I8QF14 A0A1I8MSA3 A0A3B0K1X4 A0A0M4ECF8 B4N6F4 A0A1B0A6K2 A0A1L8EF75 A0A1L8E0W6 Q2LZI3 A0A2J7Q503 B4H6N0 A0A0L0BV84 A0A1W4W4I6 A0A1L8EHX6 U5EWU3 B4IX18 B3M4I9 B4HKT0 B4QNV5 Q9VVH5 B4PJU9 B3NHU8 B4LFR8 B4KXE7 Q1W286 A0A1B6MB62 A0A1J1HWE0 A0A336LYN2 D1FQ43 A0A1W4X507 B5M0W3 A0A2M3Z047 A0A2M4A6B9 W5JLZ0 A0A2C9JTQ1 A0A224ZB31 A0A131Z597 A0A293LYV3 A0A093IPX6 A0A1L8E9I8 A0A2M4A802 D2A1S3 A0A087UF51 A0A2P6L8V1 C9W1P3 J9HFH0 A0A182G7Z1 Q7QDP2 A0A182R929 A0A3F2Z216 A0A1S4H4M9 A0A0P5TBY9 A0A084VJW1 A0A182IKK3 A0A182QGQ1 A0A094KVC3 A0A091KRL1 Q6Q9G3 K4G6V8 A0A023GEA3 A0A226E260 T1DJW3 A0A2I0T386 A0A091LNX2 A0A1B0AY58 B0WZV5 A0A182FZS5 Q09JK0 A0A0C9RZW0 Q4PM49 A0A147BVR0 B7PRC5 A0A1D2MD05 A0A3S1AQU9 A0A091FLJ2 A0A0P4YPP2 A0A091Q0X4 A0A1V4KTY2 A0A023FG99 A0A023FES7 W4ZFW5 A0A182HIW0 A0A1Q3FLY5 A0A3Q3LFP9 A0A091T0U1 A0A091S8P1 E9FT85 A0A0B7B1F4 A0A2I0LT61
PDB
4U3F
E-value=7.75371e-20,
Score=232
Ontologies
PATHWAY
GO
PANTHER
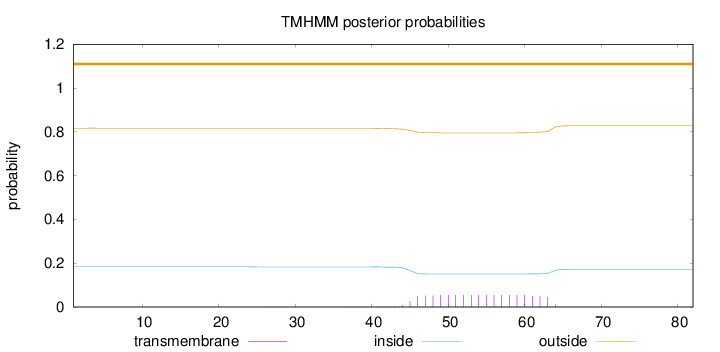
Topology
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01264
Exp number, first 60 AAs:
0.854
Total prob of N-in:
0.18329
outside
1 - 82
Population Genetic Test Statistics
Pi
123.954316
Theta
120.036819
Tajima's D
0
CLR
0.825383
CSRT
0.358982050897455
Interpretation
Uncertain
