Gene
KWMTBOMO07810 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000921
Annotation
PREDICTED:_ER_membrane_protein_complex_subunit_10_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.922
Sequence
CDS
ATGCAGCAGTTTACCTTAATATTTGTTGTACATTTAACTGCACTTTTTGCATCTACGGCCTGTATAGATTATGATGGTTTTTTGAACATGAAACTAGAACACTCTTTAAATTGCAATGATGAAAAATTTTGTCATAGAGGAAGTATAACTTTGAAGAGCATAAGAACGGGAGTTCCTATTATTGATCAAATACTATTCAACGAAAAGCATCTTGAAGCACTGAAGAAACTCGCAAATGAAGATGAATTCTATTCAATTAAAACAACAATTACAACAGGTGAAAACAGCAAAGGAACTGAATATTTATCATCGGTTAAAGCTCAAGCATTTTTGGAAAATGGTCTCTCTGATGTTATCAATGCATGGATACTACCTAATGGAGCTGTTATTGCAGTCAATTTCCAAGTTGCTAACTCTACGCAACCACTAAAGCAAGCCACTAATGAGTATAAGCTCAATTCTAAATTCTACTTAAGATATATTGAGCAAGCTCCAGTTCCCGATACTGCGTCATATATACAAAAAATGGAACGTGACAGAGAGGCACGTGAAAAGGGAGAAATGAAGGATAACAGGTCATTCTTAGCTAAATATTGGATGTATATTGTTCCAGTAGCAATATTTGTTATGATATCAGGAGCCACAAATCCAGAGGCCTCAGCACCAGCTGGACGTTAA
Protein
MQQFTLIFVVHLTALFASTACIDYDGFLNMKLEHSLNCNDEKFCHRGSITLKSIRTGVPIIDQILFNEKHLEALKKLANEDEFYSIKTTITTGENSKGTEYLSSVKAQAFLENGLSDVINAWILPNGAVIAVNFQVANSTQPLKQATNEYKLNSKFYLRYIEQAPVPDTASYIQKMERDREAREKGEMKDNRSFLAKYWMYIVPVAIFVMISGATNPEASAPAGR
Summary
Similarity
Belongs to the histone H3 family.
Uniprot
H9IUJ1
A0A2A4JG14
A0A2H1VLM8
A0A1E1WFQ7
S4PI02
A0A212EMD9
+ More
A0A194PD53 V5I7M8 A0A2J7RCM2 A0A067RIV6 A0A3B0K225 U5EW61 E0VKF8 A0A1W4WHV2 Q29F11 B4HC12 A0A0C9RIN1 A0A336KIP4 D6WD56 A0A232F031 A0A336MM68 K7IZ20 A0A1J1IQ97 A0A0M4EQ11 A0A1Y1KLK2 A0A1W4UEV5 Q17B00 A0A023EMQ7 A0A1B6DP55 B4MXQ4 A0A1I8Q7P4 A0A2A3ESX1 V9IG43 A0A026W3H6 E2BVH0 A0A1B6FPS4 E9IR87 A0A1L8EDS6 A0A1A9V301 A0A1L8EDV5 A0A1L8EE75 B3M6G9 E2AZ95 A0A195DRH9 B4KA14 A0A0L7RCM5 F4X8S4 A0A1S3D672 A0A1Q3FFG2 B0WHI3 A0A195FCQ9 A0A1B6JRP6 A0A195BAG3 A0A158NBF5 A0A182TJE8 D3TRC9 A0A1B0FKR5 T1IXV2 A0A1B6LF15 W5JQU3 B4LVT5 A0A1S4H6F5 A0A2M3ZE45 A0A182JHR1 A0A2M3Z7M9 A0A2M4BYM6 Q7PST7 C4WUD9 T1EAT9 A0A182VD09 A0A182LH83 A0A2M4APE8 A0A084VHX8 A0A151IGU9 A0A2P8YMC8 A0A1B0AR95 A0A1A9YL23 N6U1D2 B4IUM4 Q8IPT0 A0A0J9RZK6 Q6AWP4 A0A182KFN2 A0A224XVT6 E2J772 A0A182Y715 J3JU70 A0A0L0CST0 A0A0V0G379 A0A182QE93 A0A226EL24 A0A1B0D543 A0A034WMA5 B3NEB5 A0A1L8DUU1 A0A182HY40 A0A182WXG4
A0A194PD53 V5I7M8 A0A2J7RCM2 A0A067RIV6 A0A3B0K225 U5EW61 E0VKF8 A0A1W4WHV2 Q29F11 B4HC12 A0A0C9RIN1 A0A336KIP4 D6WD56 A0A232F031 A0A336MM68 K7IZ20 A0A1J1IQ97 A0A0M4EQ11 A0A1Y1KLK2 A0A1W4UEV5 Q17B00 A0A023EMQ7 A0A1B6DP55 B4MXQ4 A0A1I8Q7P4 A0A2A3ESX1 V9IG43 A0A026W3H6 E2BVH0 A0A1B6FPS4 E9IR87 A0A1L8EDS6 A0A1A9V301 A0A1L8EDV5 A0A1L8EE75 B3M6G9 E2AZ95 A0A195DRH9 B4KA14 A0A0L7RCM5 F4X8S4 A0A1S3D672 A0A1Q3FFG2 B0WHI3 A0A195FCQ9 A0A1B6JRP6 A0A195BAG3 A0A158NBF5 A0A182TJE8 D3TRC9 A0A1B0FKR5 T1IXV2 A0A1B6LF15 W5JQU3 B4LVT5 A0A1S4H6F5 A0A2M3ZE45 A0A182JHR1 A0A2M3Z7M9 A0A2M4BYM6 Q7PST7 C4WUD9 T1EAT9 A0A182VD09 A0A182LH83 A0A2M4APE8 A0A084VHX8 A0A151IGU9 A0A2P8YMC8 A0A1B0AR95 A0A1A9YL23 N6U1D2 B4IUM4 Q8IPT0 A0A0J9RZK6 Q6AWP4 A0A182KFN2 A0A224XVT6 E2J772 A0A182Y715 J3JU70 A0A0L0CST0 A0A0V0G379 A0A182QE93 A0A226EL24 A0A1B0D543 A0A034WMA5 B3NEB5 A0A1L8DUU1 A0A182HY40 A0A182WXG4
Pubmed
19121390
23622113
22118469
26354079
24845553
20566863
+ More
15632085 17994087 18362917 19820115 28648823 20075255 28004739 17510324 24945155 26483478 24508170 30249741 20798317 21282665 21719571 21347285 20353571 20920257 23761445 12364791 20966253 24438588 29403074 23537049 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25244985 22516182 26108605 25348373
15632085 17994087 18362917 19820115 28648823 20075255 28004739 17510324 24945155 26483478 24508170 30249741 20798317 21282665 21719571 21347285 20353571 20920257 23761445 12364791 20966253 24438588 29403074 23537049 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25244985 22516182 26108605 25348373
EMBL
BABH01000967
NWSH01001710
PCG70370.1
ODYU01003229
SOQ41718.1
GDQN01005256
+ More
JAT85798.1 GAIX01005605 JAA86955.1 AGBW02013857 OWR42639.1 KQ459606 KPI91256.1 GALX01006160 JAB62306.1 NEVH01005885 PNF38594.1 KK852480 KDR22968.1 OUUW01000012 SPP87373.1 GANO01003097 JAB56774.1 DS235243 EEB13874.1 CH379070 EAL29898.2 CH479275 EDW40043.1 GBYB01006756 JAG76523.1 UFQS01000317 UFQT01000317 SSX02813.1 SSX23181.1 KQ971312 EEZ98312.1 NNAY01001435 OXU23985.1 UFQS01001464 UFQT01001464 SSX11100.1 SSX30671.1 CVRI01000055 CRL01270.1 CP012526 ALC47001.1 GEZM01081920 JAV61478.1 CH477328 EAT43414.1 JXUM01086155 JXUM01095976 GAPW01003974 GAPW01003973 KQ564234 KQ563548 JAC09624.1 KXJ72550.1 KXJ73679.1 GEDC01009848 GEDC01003618 JAS27450.1 JAS33680.1 CH963876 EDW76823.1 KZ288186 PBC34797.1 JR042401 AEY59612.1 KK107453 QOIP01000004 EZA50630.1 RLU24107.1 GL450828 EFN80346.1 GECZ01017615 JAS52154.1 GL765107 EFZ16858.1 GFDG01001938 JAV16861.1 GFDG01001917 JAV16882.1 GFDG01001916 JAV16883.1 CH902618 EDV40818.1 GL444149 EFN61238.1 KQ980581 KYN15446.1 CH933806 EDW16689.1 KQ414616 KOC68501.1 GL888932 EGI57343.1 GFDL01008756 JAV26289.1 DS231936 EDS27761.1 KQ981673 KYN38193.1 GECU01005835 JAT01872.1 KQ976540 KYM81210.1 ADTU01011036 EZ423981 ADD20257.1 CCAG010023454 JH431663 GEBQ01017737 JAT22240.1 ADMH02000442 ETN66471.1 CH940650 EDW67540.1 AAAB01008816 GGFM01006007 MBW26758.1 GGFM01003788 MBW24539.1 GGFJ01008992 MBW58133.1 EAA05046.4 ABLF02029453 AK341025 BAH71509.1 GAMD01000753 JAB00838.1 GGFK01009322 MBW42643.1 ATLV01013242 KE524847 KFB37572.1 KQ977661 KYN00658.1 PYGN01000495 PSN45383.1 JXJN01002295 APGK01046695 APGK01046696 KB741066 KB632324 ENN74426.1 ERL92372.1 CM000159 CH891954 EDW95002.1 EDX00088.1 AE014296 AAN12182.2 CM002912 KMZ01049.1 BT015204 AAT94433.1 GFTR01003794 JAW12632.1 HP429290 ADN29790.1 APGK01004368 BT126783 KB735722 AEE61745.1 ENN83322.1 JRES01000064 KNC34454.1 GECL01003544 JAP02580.1 AXCN02001116 LNIX01000003 OXA57286.1 AJVK01011711 GAKP01003188 JAC55764.1 CH954178 EDV52679.1 GFDF01003895 JAV10189.1 APCN01004490
JAT85798.1 GAIX01005605 JAA86955.1 AGBW02013857 OWR42639.1 KQ459606 KPI91256.1 GALX01006160 JAB62306.1 NEVH01005885 PNF38594.1 KK852480 KDR22968.1 OUUW01000012 SPP87373.1 GANO01003097 JAB56774.1 DS235243 EEB13874.1 CH379070 EAL29898.2 CH479275 EDW40043.1 GBYB01006756 JAG76523.1 UFQS01000317 UFQT01000317 SSX02813.1 SSX23181.1 KQ971312 EEZ98312.1 NNAY01001435 OXU23985.1 UFQS01001464 UFQT01001464 SSX11100.1 SSX30671.1 CVRI01000055 CRL01270.1 CP012526 ALC47001.1 GEZM01081920 JAV61478.1 CH477328 EAT43414.1 JXUM01086155 JXUM01095976 GAPW01003974 GAPW01003973 KQ564234 KQ563548 JAC09624.1 KXJ72550.1 KXJ73679.1 GEDC01009848 GEDC01003618 JAS27450.1 JAS33680.1 CH963876 EDW76823.1 KZ288186 PBC34797.1 JR042401 AEY59612.1 KK107453 QOIP01000004 EZA50630.1 RLU24107.1 GL450828 EFN80346.1 GECZ01017615 JAS52154.1 GL765107 EFZ16858.1 GFDG01001938 JAV16861.1 GFDG01001917 JAV16882.1 GFDG01001916 JAV16883.1 CH902618 EDV40818.1 GL444149 EFN61238.1 KQ980581 KYN15446.1 CH933806 EDW16689.1 KQ414616 KOC68501.1 GL888932 EGI57343.1 GFDL01008756 JAV26289.1 DS231936 EDS27761.1 KQ981673 KYN38193.1 GECU01005835 JAT01872.1 KQ976540 KYM81210.1 ADTU01011036 EZ423981 ADD20257.1 CCAG010023454 JH431663 GEBQ01017737 JAT22240.1 ADMH02000442 ETN66471.1 CH940650 EDW67540.1 AAAB01008816 GGFM01006007 MBW26758.1 GGFM01003788 MBW24539.1 GGFJ01008992 MBW58133.1 EAA05046.4 ABLF02029453 AK341025 BAH71509.1 GAMD01000753 JAB00838.1 GGFK01009322 MBW42643.1 ATLV01013242 KE524847 KFB37572.1 KQ977661 KYN00658.1 PYGN01000495 PSN45383.1 JXJN01002295 APGK01046695 APGK01046696 KB741066 KB632324 ENN74426.1 ERL92372.1 CM000159 CH891954 EDW95002.1 EDX00088.1 AE014296 AAN12182.2 CM002912 KMZ01049.1 BT015204 AAT94433.1 GFTR01003794 JAW12632.1 HP429290 ADN29790.1 APGK01004368 BT126783 KB735722 AEE61745.1 ENN83322.1 JRES01000064 KNC34454.1 GECL01003544 JAP02580.1 AXCN02001116 LNIX01000003 OXA57286.1 AJVK01011711 GAKP01003188 JAC55764.1 CH954178 EDV52679.1 GFDF01003895 JAV10189.1 APCN01004490
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000235965
UP000027135
+ More
UP000268350 UP000009046 UP000192223 UP000001819 UP000008744 UP000007266 UP000215335 UP000002358 UP000183832 UP000092553 UP000192221 UP000008820 UP000069940 UP000249989 UP000007798 UP000095300 UP000242457 UP000053097 UP000279307 UP000008237 UP000078200 UP000007801 UP000000311 UP000078492 UP000009192 UP000053825 UP000007755 UP000079169 UP000002320 UP000078541 UP000078540 UP000005205 UP000075902 UP000092444 UP000000673 UP000008792 UP000075880 UP000007062 UP000007819 UP000075903 UP000075882 UP000030765 UP000078542 UP000245037 UP000092460 UP000092443 UP000019118 UP000030742 UP000002282 UP000000803 UP000075881 UP000076408 UP000037069 UP000075886 UP000198287 UP000092462 UP000008711 UP000075840 UP000076407
UP000268350 UP000009046 UP000192223 UP000001819 UP000008744 UP000007266 UP000215335 UP000002358 UP000183832 UP000092553 UP000192221 UP000008820 UP000069940 UP000249989 UP000007798 UP000095300 UP000242457 UP000053097 UP000279307 UP000008237 UP000078200 UP000007801 UP000000311 UP000078492 UP000009192 UP000053825 UP000007755 UP000079169 UP000002320 UP000078541 UP000078540 UP000005205 UP000075902 UP000092444 UP000000673 UP000008792 UP000075880 UP000007062 UP000007819 UP000075903 UP000075882 UP000030765 UP000078542 UP000245037 UP000092460 UP000092443 UP000019118 UP000030742 UP000002282 UP000000803 UP000075881 UP000076408 UP000037069 UP000075886 UP000198287 UP000092462 UP000008711 UP000075840 UP000076407
Interpro
Gene 3D
ProteinModelPortal
H9IUJ1
A0A2A4JG14
A0A2H1VLM8
A0A1E1WFQ7
S4PI02
A0A212EMD9
+ More
A0A194PD53 V5I7M8 A0A2J7RCM2 A0A067RIV6 A0A3B0K225 U5EW61 E0VKF8 A0A1W4WHV2 Q29F11 B4HC12 A0A0C9RIN1 A0A336KIP4 D6WD56 A0A232F031 A0A336MM68 K7IZ20 A0A1J1IQ97 A0A0M4EQ11 A0A1Y1KLK2 A0A1W4UEV5 Q17B00 A0A023EMQ7 A0A1B6DP55 B4MXQ4 A0A1I8Q7P4 A0A2A3ESX1 V9IG43 A0A026W3H6 E2BVH0 A0A1B6FPS4 E9IR87 A0A1L8EDS6 A0A1A9V301 A0A1L8EDV5 A0A1L8EE75 B3M6G9 E2AZ95 A0A195DRH9 B4KA14 A0A0L7RCM5 F4X8S4 A0A1S3D672 A0A1Q3FFG2 B0WHI3 A0A195FCQ9 A0A1B6JRP6 A0A195BAG3 A0A158NBF5 A0A182TJE8 D3TRC9 A0A1B0FKR5 T1IXV2 A0A1B6LF15 W5JQU3 B4LVT5 A0A1S4H6F5 A0A2M3ZE45 A0A182JHR1 A0A2M3Z7M9 A0A2M4BYM6 Q7PST7 C4WUD9 T1EAT9 A0A182VD09 A0A182LH83 A0A2M4APE8 A0A084VHX8 A0A151IGU9 A0A2P8YMC8 A0A1B0AR95 A0A1A9YL23 N6U1D2 B4IUM4 Q8IPT0 A0A0J9RZK6 Q6AWP4 A0A182KFN2 A0A224XVT6 E2J772 A0A182Y715 J3JU70 A0A0L0CST0 A0A0V0G379 A0A182QE93 A0A226EL24 A0A1B0D543 A0A034WMA5 B3NEB5 A0A1L8DUU1 A0A182HY40 A0A182WXG4
A0A194PD53 V5I7M8 A0A2J7RCM2 A0A067RIV6 A0A3B0K225 U5EW61 E0VKF8 A0A1W4WHV2 Q29F11 B4HC12 A0A0C9RIN1 A0A336KIP4 D6WD56 A0A232F031 A0A336MM68 K7IZ20 A0A1J1IQ97 A0A0M4EQ11 A0A1Y1KLK2 A0A1W4UEV5 Q17B00 A0A023EMQ7 A0A1B6DP55 B4MXQ4 A0A1I8Q7P4 A0A2A3ESX1 V9IG43 A0A026W3H6 E2BVH0 A0A1B6FPS4 E9IR87 A0A1L8EDS6 A0A1A9V301 A0A1L8EDV5 A0A1L8EE75 B3M6G9 E2AZ95 A0A195DRH9 B4KA14 A0A0L7RCM5 F4X8S4 A0A1S3D672 A0A1Q3FFG2 B0WHI3 A0A195FCQ9 A0A1B6JRP6 A0A195BAG3 A0A158NBF5 A0A182TJE8 D3TRC9 A0A1B0FKR5 T1IXV2 A0A1B6LF15 W5JQU3 B4LVT5 A0A1S4H6F5 A0A2M3ZE45 A0A182JHR1 A0A2M3Z7M9 A0A2M4BYM6 Q7PST7 C4WUD9 T1EAT9 A0A182VD09 A0A182LH83 A0A2M4APE8 A0A084VHX8 A0A151IGU9 A0A2P8YMC8 A0A1B0AR95 A0A1A9YL23 N6U1D2 B4IUM4 Q8IPT0 A0A0J9RZK6 Q6AWP4 A0A182KFN2 A0A224XVT6 E2J772 A0A182Y715 J3JU70 A0A0L0CST0 A0A0V0G379 A0A182QE93 A0A226EL24 A0A1B0D543 A0A034WMA5 B3NEB5 A0A1L8DUU1 A0A182HY40 A0A182WXG4
Ontologies
GO
PANTHER
Topology
Subcellular location
Chromosome
Nucleus
Nucleus
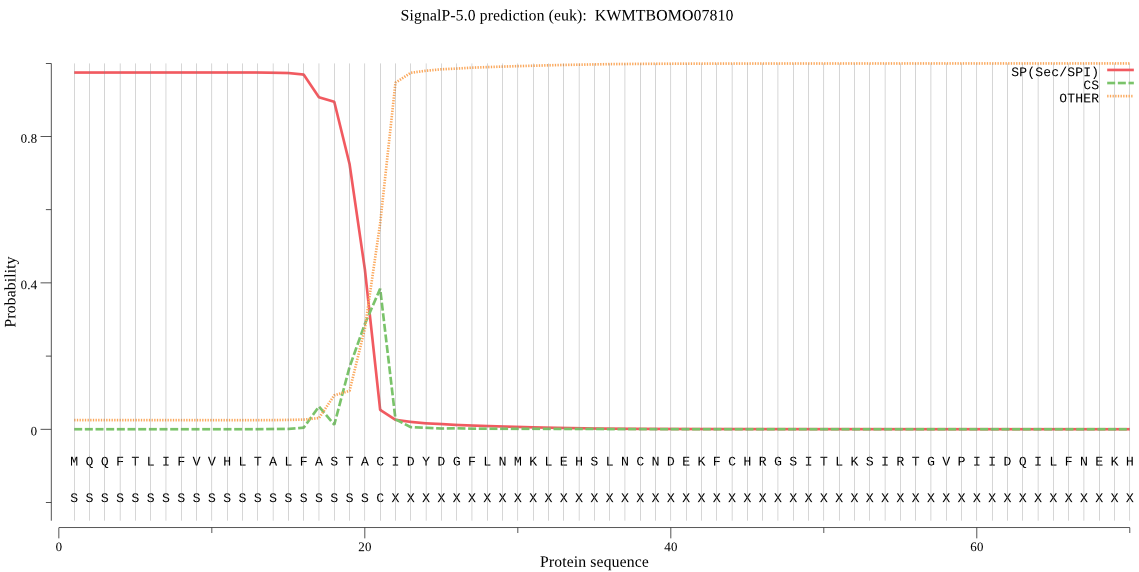
SignalP
Position: 1 - 21,
Likelihood: 0.974871
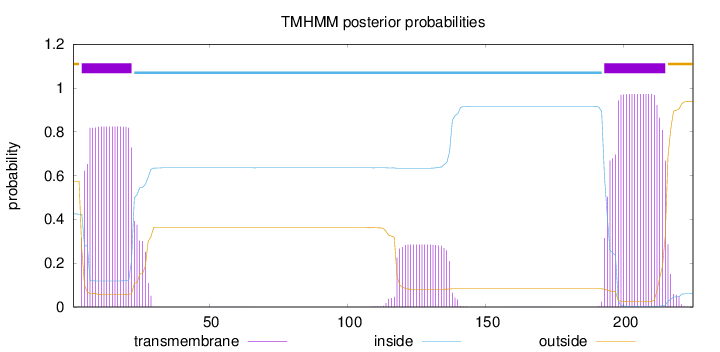
Length:
225
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
42.57066
Exp number, first 60 AAs:
16.44286
Total prob of N-in:
0.42578
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 22
inside
23 - 192
TMhelix
193 - 215
outside
216 - 225
Population Genetic Test Statistics
Pi
175.537149
Theta
144.483618
Tajima's D
0.559997
CLR
0.170813
CSRT
0.529123543822809
Interpretation
Uncertain
