Gene
KWMTBOMO07808
Pre Gene Modal
BGIBMGA001224
Annotation
PREDICTED:_delta-sarcoglycan-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.286 PlasmaMembrane Reliability : 1.939
Sequence
CDS
ATGTGTGTGTGGGGTGTGAAACAGCAGAACGGCGGTATCGACGACCTGAGGATACCGTACGCCGACAAGATCACCCCGGAGCCAATCTTGCACAACAATGCGGGCAGGGACACAAAGCCAGACTCGATCAGAAACAGTTATAATAGCCAATTCAAAGTTGGCATCTACGGTTGGAGGAAGAAATGCCTGTATATACTTGTCATTGCGCTTATGTTTATGATGATCATTAATCTAGCTCTCACGCTTTGGGTTCTCAAAGTTTTGGATTTTAATTCGGAAGGAATGGGTCAGCTCAGAATAGTTCCTGGAGGATTGCAGCTACTCGGGCAAGCATTAGTACTCGATTCTCTATTTACATCAAGTATCAAATCTCGGAGAGGTCAACCGATTTCCATCGAATCTACCAGAAACTTCTCAGTGTCAACTCGAGATGCACAGGGATTACTGCAAAGTAGATTATTCTTAGGTCATGACCGTTTAGAACTCAACGTTGGTCGCTTTGAAGTGAGAGATTCAAGAGGAGCGTTGCTACTGGGAGCCGGTCAAGATTCCGTCACCATAGGAGCGGATACACTAACGGTTTCCAGTTCAGCTGGAGTGACACTGAATACCGCAGTCCAGACTTCACTTGTAAAGGCACCCCCAGCGAAACCATTAGTGCTGGAGTCTCCTACAAGATCATTAGAAATGCATGCAGCCCAAAACATAGCTTTGGAGTCAAGAGCTGGCGACATAAGTGCGAGCTGCCTCACAACCTTCAGACTGAGATCCATAGCCGGATCGATACGTTTAGACGCACCTAGTATTTACATGCCGAAATTGAAATCGGCTCTACCATTGCCACCATCAGCACAGCACCCGCACGATCCCCATCACCAGAACATTTATCAGTTGTGTGCATGTGCAAACGGGAAACTATTCCTAGCACCACCGCATGGGATCTGCGCGGCCAGGGAGGAGAGCCTCATCTGTCGATGA
Protein
MCVWGVKQQNGGIDDLRIPYADKITPEPILHNNAGRDTKPDSIRNSYNSQFKVGIYGWRKKCLYILVIALMFMMIINLALTLWVLKVLDFNSEGMGQLRIVPGGLQLLGQALVLDSLFTSSIKSRRGQPISIESTRNFSVSTRDAQGLLQSRLFLGHDRLELNVGRFEVRDSRGALLLGAGQDSVTIGADTLTVSSSAGVTLNTAVQTSLVKAPPAKPLVLESPTRSLEMHAAQNIALESRAGDISASCLTTFRLRSIAGSIRLDAPSIYMPKLKSALPLPPSAQHPHDPHHQNIYQLCACANGKLFLAPPHGICAAREESLICR
Summary
Uniprot
H9IVE4
A0A3S2TDT4
A0A2A4IUV5
A0A2H1V8S0
A0A194RHG4
A0A212EMB7
+ More
A0A194PD49 A0A0L7LSP2 A0A2J7Q101 B0XJN9 A0A067R3F3 A0A182VVP4 A0A182IZG3 A0A1L8E5F9 A0A1L8E5D7 A0A182XER6 A0A182HIP2 A0A1W7R920 A0A182RZ34 A0A182V7D4 A0A182PKX1 A0A182Q3J7 A0A139WJ89 A0A182MER8 A0A336MWC4 A0A084VJ08 A0A336LCK3 A0A0A1WDX2 Q7QEN3 A0A034VT16 A0A1Y1LEB4 A0A3B0KJR1 N6TUN7 B4GTI4 W8B695 Q29J24 B4PY54 B3P9L3 A0A0L7QMM1 O76892 E2C091 A0A1I8NEH9 B4NEE8 B4JLW6 A0A1W4VFZ2 A0A0L0CP83 N6TR09 A0A154P8M7 B4MGX9 A0A1A9UZQ8 A0A026W1R6 B3MYM0 A0A1A9YI61 A0A1B0AHU2 A0A088ALA6 B4L778 A0A1A9WCH3 A0A1B0B2C5 A0A2P8YTJ9 A0A087T563 A0A1B0GEF9 C0P8L9 E9HAK5 A0A0M4F878 A0A1D2N2P5 A0A1I8NX46 A0A023ENJ0 U4UFT1 N6UMJ5 A0A0P6JZZ8 A0A151I3V0 A0A226E8B2 A0A023F0F5 A0A182HEH8 A0A0K8SBJ4 Q178M3 A0A0C9R9I4 A0A151WYC4 Q16EP8 A0A0A9X6V5 A0A182UF70 A0A1B0DEM4 A0A0C9QMS5 A0A0C9PTZ8 A0A0P6AB81 A0A182FJE2 A0A3B4GNL2 A0A3Q3C2E3 A0A3Q4GBJ9 A0A3P9AYR9 G3Q3V9 A0A3P8NBM6 I3KGW7 R7TE37 A0A3P9IL75 A0A3Q0RHL2 A0A3P8S0Y3
A0A194PD49 A0A0L7LSP2 A0A2J7Q101 B0XJN9 A0A067R3F3 A0A182VVP4 A0A182IZG3 A0A1L8E5F9 A0A1L8E5D7 A0A182XER6 A0A182HIP2 A0A1W7R920 A0A182RZ34 A0A182V7D4 A0A182PKX1 A0A182Q3J7 A0A139WJ89 A0A182MER8 A0A336MWC4 A0A084VJ08 A0A336LCK3 A0A0A1WDX2 Q7QEN3 A0A034VT16 A0A1Y1LEB4 A0A3B0KJR1 N6TUN7 B4GTI4 W8B695 Q29J24 B4PY54 B3P9L3 A0A0L7QMM1 O76892 E2C091 A0A1I8NEH9 B4NEE8 B4JLW6 A0A1W4VFZ2 A0A0L0CP83 N6TR09 A0A154P8M7 B4MGX9 A0A1A9UZQ8 A0A026W1R6 B3MYM0 A0A1A9YI61 A0A1B0AHU2 A0A088ALA6 B4L778 A0A1A9WCH3 A0A1B0B2C5 A0A2P8YTJ9 A0A087T563 A0A1B0GEF9 C0P8L9 E9HAK5 A0A0M4F878 A0A1D2N2P5 A0A1I8NX46 A0A023ENJ0 U4UFT1 N6UMJ5 A0A0P6JZZ8 A0A151I3V0 A0A226E8B2 A0A023F0F5 A0A182HEH8 A0A0K8SBJ4 Q178M3 A0A0C9R9I4 A0A151WYC4 Q16EP8 A0A0A9X6V5 A0A182UF70 A0A1B0DEM4 A0A0C9QMS5 A0A0C9PTZ8 A0A0P6AB81 A0A182FJE2 A0A3B4GNL2 A0A3Q3C2E3 A0A3Q4GBJ9 A0A3P9AYR9 G3Q3V9 A0A3P8NBM6 I3KGW7 R7TE37 A0A3P9IL75 A0A3Q0RHL2 A0A3P8S0Y3
Pubmed
19121390
26354079
22118469
26227816
24845553
18362917
+ More
19820115 24438588 25830018 12364791 25348373 28004739 23537049 17994087 24495485 15632085 17550304 18057021 11018515 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 25315136 26108605 24508170 29403074 21292972 27289101 24945155 25474469 26483478 17510324 25401762 25186727 23254933 17554307
19820115 24438588 25830018 12364791 25348373 28004739 23537049 17994087 24495485 15632085 17550304 18057021 11018515 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 25315136 26108605 24508170 29403074 21292972 27289101 24945155 25474469 26483478 17510324 25401762 25186727 23254933 17554307
EMBL
BABH01000977
RSAL01000276
RVE43105.1
NWSH01006006
PCG63757.1
ODYU01001018
+ More
SOQ36654.1 KQ460205 KPJ16879.1 AGBW02013857 OWR42634.1 KQ459606 KPI91251.1 JTDY01000166 KOB78485.1 NEVH01019960 PNF22270.1 DS233570 EDS30558.1 KK852782 KDR16618.1 GFDF01000146 JAV13938.1 GFDF01000342 JAV13742.1 APCN01004029 APCN01004030 APCN01004031 GEHC01000004 JAV47641.1 AXCN02001221 KQ971338 KYB27911.1 AXCM01009022 UFQT01002640 SSX33831.1 ATLV01013422 KE524855 KFB37952.1 UFQS01002721 UFQT01002721 SSX14591.1 SSX33989.1 GBXI01017622 JAC96669.1 AAAB01008846 EAA06160.5 GAKP01013977 JAC44975.1 GEZM01060161 JAV71231.1 OUUW01000016 SPP88810.1 APGK01017892 APGK01017893 KB740057 ENN81788.1 CH479190 EDW25854.1 GAMC01012433 JAB94122.1 CH379063 EAL32478.2 CM000162 EDX00927.1 CH954183 EDV45509.1 KQS25987.1 KQ414889 KOC59809.1 AE014298 AF277393 BT064640 AL030994 AAF45663.1 AAG17402.1 ACN29337.1 ACZ95176.2 ACZ95177.1 ACZ95178.1 CAA19736.1 GL451753 EFN78643.1 CH964239 EDW82117.2 CH916371 EDV91727.1 JRES01000125 KNC33987.1 APGK01008818 APGK01008819 KB737790 ENN82924.1 KQ434827 KZC07480.1 CH940757 EDW71529.1 KK107488 EZA50010.1 CH902632 EDV32714.1 CH933812 EDW06224.2 JXJN01007595 JXJN01007596 JXJN01007597 PYGN01000367 PSN47579.1 KK113457 KFM60252.1 CCAG010012243 CCAG010012244 CCAG010012245 BT064638 ACN29335.1 GL732612 EFX71196.1 CP012528 ALC48182.1 LJIJ01000275 ODM99510.1 GAPW01003624 JAC09974.1 KB632278 ERL91228.1 APGK01008440 APGK01008441 APGK01008812 APGK01008813 KB737789 KB737692 ENN82925.1 ENN82960.1 GDIQ01001237 LRGB01000248 JAN93500.1 KZS20050.1 KQ976463 KYM84598.1 LNIX01000006 OXA53247.1 GBBI01003775 JAC14937.1 JXUM01035407 KQ561058 KXJ79834.1 GBRD01015154 JAG50672.1 CH477361 EAT42667.1 GBYB01004795 GBYB01004796 JAG74562.1 JAG74563.1 KQ982656 KYQ52701.1 CH478624 EAT32707.1 GBHO01044142 GBHO01027147 GBHO01027146 GBHO01017072 GBHO01017071 GBHO01017069 GBRD01015155 GBRD01015153 JAF99461.1 JAG16457.1 JAG16458.1 JAG26532.1 JAG26533.1 JAG26535.1 JAG50671.1 AJVK01005661 GBYB01004794 JAG74561.1 GBYB01004793 JAG74560.1 GDIP01031960 JAM71755.1 AERX01023408 AERX01023409 AERX01023410 AERX01023411 AERX01023412 AMQN01003279 KB311417 ELT89316.1
SOQ36654.1 KQ460205 KPJ16879.1 AGBW02013857 OWR42634.1 KQ459606 KPI91251.1 JTDY01000166 KOB78485.1 NEVH01019960 PNF22270.1 DS233570 EDS30558.1 KK852782 KDR16618.1 GFDF01000146 JAV13938.1 GFDF01000342 JAV13742.1 APCN01004029 APCN01004030 APCN01004031 GEHC01000004 JAV47641.1 AXCN02001221 KQ971338 KYB27911.1 AXCM01009022 UFQT01002640 SSX33831.1 ATLV01013422 KE524855 KFB37952.1 UFQS01002721 UFQT01002721 SSX14591.1 SSX33989.1 GBXI01017622 JAC96669.1 AAAB01008846 EAA06160.5 GAKP01013977 JAC44975.1 GEZM01060161 JAV71231.1 OUUW01000016 SPP88810.1 APGK01017892 APGK01017893 KB740057 ENN81788.1 CH479190 EDW25854.1 GAMC01012433 JAB94122.1 CH379063 EAL32478.2 CM000162 EDX00927.1 CH954183 EDV45509.1 KQS25987.1 KQ414889 KOC59809.1 AE014298 AF277393 BT064640 AL030994 AAF45663.1 AAG17402.1 ACN29337.1 ACZ95176.2 ACZ95177.1 ACZ95178.1 CAA19736.1 GL451753 EFN78643.1 CH964239 EDW82117.2 CH916371 EDV91727.1 JRES01000125 KNC33987.1 APGK01008818 APGK01008819 KB737790 ENN82924.1 KQ434827 KZC07480.1 CH940757 EDW71529.1 KK107488 EZA50010.1 CH902632 EDV32714.1 CH933812 EDW06224.2 JXJN01007595 JXJN01007596 JXJN01007597 PYGN01000367 PSN47579.1 KK113457 KFM60252.1 CCAG010012243 CCAG010012244 CCAG010012245 BT064638 ACN29335.1 GL732612 EFX71196.1 CP012528 ALC48182.1 LJIJ01000275 ODM99510.1 GAPW01003624 JAC09974.1 KB632278 ERL91228.1 APGK01008440 APGK01008441 APGK01008812 APGK01008813 KB737789 KB737692 ENN82925.1 ENN82960.1 GDIQ01001237 LRGB01000248 JAN93500.1 KZS20050.1 KQ976463 KYM84598.1 LNIX01000006 OXA53247.1 GBBI01003775 JAC14937.1 JXUM01035407 KQ561058 KXJ79834.1 GBRD01015154 JAG50672.1 CH477361 EAT42667.1 GBYB01004795 GBYB01004796 JAG74562.1 JAG74563.1 KQ982656 KYQ52701.1 CH478624 EAT32707.1 GBHO01044142 GBHO01027147 GBHO01027146 GBHO01017072 GBHO01017071 GBHO01017069 GBRD01015155 GBRD01015153 JAF99461.1 JAG16457.1 JAG16458.1 JAG26532.1 JAG26533.1 JAG26535.1 JAG50671.1 AJVK01005661 GBYB01004794 JAG74561.1 GBYB01004793 JAG74560.1 GDIP01031960 JAM71755.1 AERX01023408 AERX01023409 AERX01023410 AERX01023411 AERX01023412 AMQN01003279 KB311417 ELT89316.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000007151
UP000053268
+ More
UP000037510 UP000235965 UP000002320 UP000027135 UP000075920 UP000075880 UP000076407 UP000075840 UP000075900 UP000075903 UP000075885 UP000075886 UP000007266 UP000075883 UP000030765 UP000007062 UP000268350 UP000019118 UP000008744 UP000001819 UP000002282 UP000008711 UP000053825 UP000000803 UP000008237 UP000095301 UP000007798 UP000001070 UP000192221 UP000037069 UP000076502 UP000008792 UP000078200 UP000053097 UP000007801 UP000092443 UP000092445 UP000005203 UP000009192 UP000091820 UP000092460 UP000245037 UP000054359 UP000092444 UP000000305 UP000092553 UP000094527 UP000095300 UP000030742 UP000076858 UP000078540 UP000198287 UP000069940 UP000249989 UP000008820 UP000075809 UP000075902 UP000092462 UP000069272 UP000261460 UP000264840 UP000261580 UP000265160 UP000007635 UP000265100 UP000005207 UP000014760 UP000265180 UP000265200 UP000261340 UP000265080
UP000037510 UP000235965 UP000002320 UP000027135 UP000075920 UP000075880 UP000076407 UP000075840 UP000075900 UP000075903 UP000075885 UP000075886 UP000007266 UP000075883 UP000030765 UP000007062 UP000268350 UP000019118 UP000008744 UP000001819 UP000002282 UP000008711 UP000053825 UP000000803 UP000008237 UP000095301 UP000007798 UP000001070 UP000192221 UP000037069 UP000076502 UP000008792 UP000078200 UP000053097 UP000007801 UP000092443 UP000092445 UP000005203 UP000009192 UP000091820 UP000092460 UP000245037 UP000054359 UP000092444 UP000000305 UP000092553 UP000094527 UP000095300 UP000030742 UP000076858 UP000078540 UP000198287 UP000069940 UP000249989 UP000008820 UP000075809 UP000075902 UP000092462 UP000069272 UP000261460 UP000264840 UP000261580 UP000265160 UP000007635 UP000265100 UP000005207 UP000014760 UP000265180 UP000265200 UP000261340 UP000265080
Pfam
PF04790 Sarcoglycan_1
ProteinModelPortal
H9IVE4
A0A3S2TDT4
A0A2A4IUV5
A0A2H1V8S0
A0A194RHG4
A0A212EMB7
+ More
A0A194PD49 A0A0L7LSP2 A0A2J7Q101 B0XJN9 A0A067R3F3 A0A182VVP4 A0A182IZG3 A0A1L8E5F9 A0A1L8E5D7 A0A182XER6 A0A182HIP2 A0A1W7R920 A0A182RZ34 A0A182V7D4 A0A182PKX1 A0A182Q3J7 A0A139WJ89 A0A182MER8 A0A336MWC4 A0A084VJ08 A0A336LCK3 A0A0A1WDX2 Q7QEN3 A0A034VT16 A0A1Y1LEB4 A0A3B0KJR1 N6TUN7 B4GTI4 W8B695 Q29J24 B4PY54 B3P9L3 A0A0L7QMM1 O76892 E2C091 A0A1I8NEH9 B4NEE8 B4JLW6 A0A1W4VFZ2 A0A0L0CP83 N6TR09 A0A154P8M7 B4MGX9 A0A1A9UZQ8 A0A026W1R6 B3MYM0 A0A1A9YI61 A0A1B0AHU2 A0A088ALA6 B4L778 A0A1A9WCH3 A0A1B0B2C5 A0A2P8YTJ9 A0A087T563 A0A1B0GEF9 C0P8L9 E9HAK5 A0A0M4F878 A0A1D2N2P5 A0A1I8NX46 A0A023ENJ0 U4UFT1 N6UMJ5 A0A0P6JZZ8 A0A151I3V0 A0A226E8B2 A0A023F0F5 A0A182HEH8 A0A0K8SBJ4 Q178M3 A0A0C9R9I4 A0A151WYC4 Q16EP8 A0A0A9X6V5 A0A182UF70 A0A1B0DEM4 A0A0C9QMS5 A0A0C9PTZ8 A0A0P6AB81 A0A182FJE2 A0A3B4GNL2 A0A3Q3C2E3 A0A3Q4GBJ9 A0A3P9AYR9 G3Q3V9 A0A3P8NBM6 I3KGW7 R7TE37 A0A3P9IL75 A0A3Q0RHL2 A0A3P8S0Y3
A0A194PD49 A0A0L7LSP2 A0A2J7Q101 B0XJN9 A0A067R3F3 A0A182VVP4 A0A182IZG3 A0A1L8E5F9 A0A1L8E5D7 A0A182XER6 A0A182HIP2 A0A1W7R920 A0A182RZ34 A0A182V7D4 A0A182PKX1 A0A182Q3J7 A0A139WJ89 A0A182MER8 A0A336MWC4 A0A084VJ08 A0A336LCK3 A0A0A1WDX2 Q7QEN3 A0A034VT16 A0A1Y1LEB4 A0A3B0KJR1 N6TUN7 B4GTI4 W8B695 Q29J24 B4PY54 B3P9L3 A0A0L7QMM1 O76892 E2C091 A0A1I8NEH9 B4NEE8 B4JLW6 A0A1W4VFZ2 A0A0L0CP83 N6TR09 A0A154P8M7 B4MGX9 A0A1A9UZQ8 A0A026W1R6 B3MYM0 A0A1A9YI61 A0A1B0AHU2 A0A088ALA6 B4L778 A0A1A9WCH3 A0A1B0B2C5 A0A2P8YTJ9 A0A087T563 A0A1B0GEF9 C0P8L9 E9HAK5 A0A0M4F878 A0A1D2N2P5 A0A1I8NX46 A0A023ENJ0 U4UFT1 N6UMJ5 A0A0P6JZZ8 A0A151I3V0 A0A226E8B2 A0A023F0F5 A0A182HEH8 A0A0K8SBJ4 Q178M3 A0A0C9R9I4 A0A151WYC4 Q16EP8 A0A0A9X6V5 A0A182UF70 A0A1B0DEM4 A0A0C9QMS5 A0A0C9PTZ8 A0A0P6AB81 A0A182FJE2 A0A3B4GNL2 A0A3Q3C2E3 A0A3Q4GBJ9 A0A3P9AYR9 G3Q3V9 A0A3P8NBM6 I3KGW7 R7TE37 A0A3P9IL75 A0A3Q0RHL2 A0A3P8S0Y3
Ontologies
GO
PANTHER
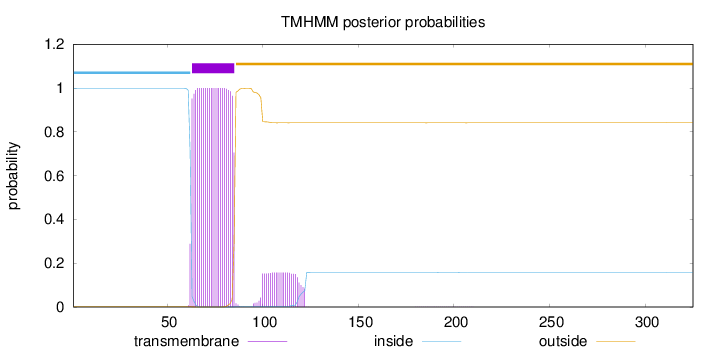
Topology
Length:
325
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
26.3334499999999
Exp number, first 60 AAs:
0.00065
Total prob of N-in:
0.99772
inside
1 - 62
TMhelix
63 - 85
outside
86 - 325
Population Genetic Test Statistics
Pi
149.878602
Theta
178.825095
Tajima's D
-0.561763
CLR
0.859767
CSRT
0.228538573071346
Interpretation
Uncertain
