Gene
KWMTBOMO07803
Pre Gene Modal
BGIBMGA001230
Annotation
PREDICTED:_otopetrin-2-like_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.905
Sequence
CDS
ATGAGTCCGGCCAATCGCAGCCCACTGTGCGAGGCGAACGAATCTCATCCCGGCGTCACGATAACAGACATGGACACGGTGCATGAAGAAAACGATAATGACAGTGATGAATCGTCGTACTCAGATCTAGGTCGGCTCCGAGAGCGTACTAAAGCGATGCTCGGAACCCCTGCAGATCCGCTGATCCAGCATTCACACTCGCAGCCAAACAGCAGGAGAATGTCCTCGGATATCGAAACTAAACAGCTAGCAGCCAATCTCAATACAAGACGTCCATCCGCAATTATAGCAGCTTTCCGACGACCATCGCAAGCATTAGCGCTATGTGCTGCCACACAACGTCGCTTCCTATCTGAATTGACACCGCATTCGAGGGCATCCATGGCATCTACAATACATGAGACAGCACCATCGGCTGCGGAGATTTTAGGGCATGAAGCTTTAAGCAAAGCGTTGTCCGCTTTGTATGCAAAGTTGCTCGTTGTGTTGGGCTTAGCTTTTCCAGTCACTGAAGTGATAGCTAAGGGCGTGCCAGACGGATATTATCAGGGATTTTATTTGTACCTGTATTCAGTATCAGTTCTATTTGTCGTTTTTCAATACGCAAATATAATGCGACAGAAGGCGGTGACTATGATCATAAGTAACTACGGTCCAGATGGCGAAAAAGAAGACAAAGGTAGAACAACTCCAGCAGAAAAATTAAAAATAAAAGAAACTATCTCTCGATACGGAAGCTTTTATTTGAGACTTGGTGCTATCGCATTCGGTATAGGTTCGATGGTGTACAGTGGCTTAGAATTTGGGGAATATTTCGAGATGACAGACCAATGCCGTTCAATACTGGCAGTCATTACGCCAGCCCTAAGAATGACTTTAACTTTAGCACAAATGCAATTTATATTCCTCACGAACAAAGACATAGAGTTGGGAAGCCACAAGATGATCCAAAGATTCGGTTTCATGCACATGTTGGCAACTAATCTATGCGAATGGCTATACGTGCTAGTTGAGGAAACTAAACATGAAATACATCATTTGGAGCACGCCATGCTGTCTCCAGAAAATACTACTCATAACGTTACGGAAGAGATATCCTGTCGAAGATCAAATATAATGGGTGCTCTATTACAACGTGCCTCACCATTTCTCTTTCCTTGTACGATAGAATATTCACTTATCTGTGCGGTCATTTTTTATGAAATGTGGAAAGAGATAAAATGTACACCGGAGGATTTTGAAAAAAAATTCCACGTCCCCAAAAATAAATCACGAAACAACTCCGTTACGCCGGAGAAAATTTTGCAGCAGTTTGCTGGTATGCTTGGTGTGAGCTCTCCACACGGCAGTCGCACCGCCCTACATCACTTCTCTGTGGATTGTTCGCACGCACACCGCGGACTCTCCGCTGGTGTTCTCATCATCGTCCTCACCATTATATCGCTCATCATGTTCTTCGTATTAGCTAACAACCCAGATACTGTAATCGATGCTATATTTGAAGTTAATGTTTGTGAGTTGGTTTTATACACGATGACATTAATAGCTGCCGGTGCCGCATTATTACAAATGCGTGCTCTTCCTTATAAAAGGAAATCTCAAGCTGTATTAGGTCTAGACACATCACTACTGATCTTGGCTCAGACCGGTATGTTCGTGTACTGCATGTTCAGTTTGATCGGATGTCATCATACTATGAACAGCAATAACAGTTCCGGGTATACTGGTTTCTTCTCAGAGTTCTTATCTCTCATACAGACAATACTGCAGTCTTTATTTATCATCGATGCTTGGTGGCGGCGATGCAGCAATTCAGAGCAACGGCGAGCGAAGCCGGGACGACAATTAGTCACATTCTTGCTCGTGGCAAACATGGCGATGTGGACCATCAATACACTAGAAAAGAATAGAGCCGAGTTTCGTCCAGCACATCTGGCTTTCTACGGACCTTGGGCGTGGACAATAATCACGCATGTTTCGATGCCTCTCGCCATTTTCTACCGCTTCCACTCCACTATTTGTCTATTCGAGATATGGAAGAATTGTTTCAAAAACAAACCAATATACTTAAATGTTTAG
Protein
MSPANRSPLCEANESHPGVTITDMDTVHEENDNDSDESSYSDLGRLRERTKAMLGTPADPLIQHSHSQPNSRRMSSDIETKQLAANLNTRRPSAIIAAFRRPSQALALCAATQRRFLSELTPHSRASMASTIHETAPSAAEILGHEALSKALSALYAKLLVVLGLAFPVTEVIAKGVPDGYYQGFYLYLYSVSVLFVVFQYANIMRQKAVTMIISNYGPDGEKEDKGRTTPAEKLKIKETISRYGSFYLRLGAIAFGIGSMVYSGLEFGEYFEMTDQCRSILAVITPALRMTLTLAQMQFIFLTNKDIELGSHKMIQRFGFMHMLATNLCEWLYVLVEETKHEIHHLEHAMLSPENTTHNVTEEISCRRSNIMGALLQRASPFLFPCTIEYSLICAVIFYEMWKEIKCTPEDFEKKFHVPKNKSRNNSVTPEKILQQFAGMLGVSSPHGSRTALHHFSVDCSHAHRGLSAGVLIIVLTIISLIMFFVLANNPDTVIDAIFEVNVCELVLYTMTLIAAGAALLQMRALPYKRKSQAVLGLDTSLLILAQTGMFVYCMFSLIGCHHTMNSNNSSGYTGFFSEFLSLIQTILQSLFIIDAWWRRCSNSEQRRAKPGRQLVTFLLVANMAMWTINTLEKNRAEFRPAHLAFYGPWAWTIITHVSMPLAIFYRFHSTICLFEIWKNCFKNKPIYLNV
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
A0A2H1VCH1
A0A194RHF9
A0A194PJI1
A0A1E1W187
A0A212EMC9
A0A3S2KYZ8
+ More
A0A0L7KTX5 V5G3P9 D2A3K9 N6TQA5 A0A1B0DPH1 A0A1Y1LL30 U4UBC1 A0A1L8D8X6 A0A1L8D8V6 Q0IFL7 A0A1L8D8X9 B0X0Q3 A0A182PKX5 A0A0K8TQR5 A0A1J1HXL1 A0A182MBX3 Q7QEN7 A0A182XES1 A0A182HIN8 A0A182JKA1 W5J828 A0A182FJD8 A0A084VJ21 A0A1B0ETG4 A0A182YCN0 A0A182Q0E2 A0A182RZ31 A0A182U4E0 A0A336LVG2 A0A336KML1 A0A182VVP7 A0A182N028 E0VCH7 B0X0Q2 A0A336M5E8 A0A1B6C2N4 A0A1S4F8C6 Q0IFL6 A0A067R125 A0A182HB61 A0A2P8YTK7 A0A034W8D4 A0A0K8V3B4 A0A3B0JIZ8 A0A1I8NPZ8 A0A0R3P033 A0A2J7R2L2 A0A3B0K850 A0A3B0KF18 A0A146LN45 A0A1I8M6Y2 A0A1I8M6Y3 A0A088AUK5 A0A0K8VFW9 A0A1W4V6L7 A0A1W4UU99 A0A2R7WDC1 A0A3B0JIL3 A0A3B0K9A0 E2BVD7 B4GSF2 A0A1A9WCG6 Q8IQ02 A0A026W3F3 Q29L36 A0A0A1X4K0 V9IDZ6 X2J8I0 A0A0R1DJ26 A0A0Q5W8X6 A0A0L0BY36 A0A1W4UUN8 A0A0A1XQ32 A0A0J9QUY9 F4X8Q0 A0A1B6CS77 A0A0J9QUT9 A0A3B0K2U2 A0A1B0FRD1 A0A154NXD9 A0A232EUU7 A0A0P9A708 B4NEF1 A0A0L7R7U6 H9IVH9 A0A2A4JFV7 X2JD17 Q9VQN3 A0A1A9YQR1 W8BVT8 A0A1B0B143 A0A1A9UZS0
A0A0L7KTX5 V5G3P9 D2A3K9 N6TQA5 A0A1B0DPH1 A0A1Y1LL30 U4UBC1 A0A1L8D8X6 A0A1L8D8V6 Q0IFL7 A0A1L8D8X9 B0X0Q3 A0A182PKX5 A0A0K8TQR5 A0A1J1HXL1 A0A182MBX3 Q7QEN7 A0A182XES1 A0A182HIN8 A0A182JKA1 W5J828 A0A182FJD8 A0A084VJ21 A0A1B0ETG4 A0A182YCN0 A0A182Q0E2 A0A182RZ31 A0A182U4E0 A0A336LVG2 A0A336KML1 A0A182VVP7 A0A182N028 E0VCH7 B0X0Q2 A0A336M5E8 A0A1B6C2N4 A0A1S4F8C6 Q0IFL6 A0A067R125 A0A182HB61 A0A2P8YTK7 A0A034W8D4 A0A0K8V3B4 A0A3B0JIZ8 A0A1I8NPZ8 A0A0R3P033 A0A2J7R2L2 A0A3B0K850 A0A3B0KF18 A0A146LN45 A0A1I8M6Y2 A0A1I8M6Y3 A0A088AUK5 A0A0K8VFW9 A0A1W4V6L7 A0A1W4UU99 A0A2R7WDC1 A0A3B0JIL3 A0A3B0K9A0 E2BVD7 B4GSF2 A0A1A9WCG6 Q8IQ02 A0A026W3F3 Q29L36 A0A0A1X4K0 V9IDZ6 X2J8I0 A0A0R1DJ26 A0A0Q5W8X6 A0A0L0BY36 A0A1W4UUN8 A0A0A1XQ32 A0A0J9QUY9 F4X8Q0 A0A1B6CS77 A0A0J9QUT9 A0A3B0K2U2 A0A1B0FRD1 A0A154NXD9 A0A232EUU7 A0A0P9A708 B4NEF1 A0A0L7R7U6 H9IVH9 A0A2A4JFV7 X2JD17 Q9VQN3 A0A1A9YQR1 W8BVT8 A0A1B0B143 A0A1A9UZS0
Pubmed
26354079
22118469
26227816
18362917
19820115
23537049
+ More
28004739 17510324 26369729 12364791 20920257 23761445 24438588 25244985 20566863 24845553 26483478 29403074 25348373 15632085 17994087 26823975 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 25830018 17550304 26108605 22936249 21719571 28648823 19121390 24495485
28004739 17510324 26369729 12364791 20920257 23761445 24438588 25244985 20566863 24845553 26483478 29403074 25348373 15632085 17994087 26823975 25315136 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 30249741 25830018 17550304 26108605 22936249 21719571 28648823 19121390 24495485
EMBL
ODYU01001821
SOQ38548.1
KQ460205
KPJ16874.1
KQ459606
KPI91245.1
+ More
GDQN01010297 GDQN01004622 JAT80757.1 JAT86432.1 AGBW02013857 OWR42629.1 RSAL01000413 RVE41839.1 JTDY01005896 KOB66504.1 GALX01003816 JAB64650.1 KQ971338 EFA01890.1 APGK01010729 KB738283 ENN82679.1 AJVK01018277 AJVK01018278 GEZM01054482 JAV73621.1 KB632278 ERL91229.1 GFDF01011173 JAV02911.1 GFDF01011191 JAV02893.1 CH477312 EAT43886.1 GFDF01011172 JAV02912.1 DS232243 EDS38295.1 GDAI01001110 JAI16493.1 CVRI01000035 CRK92831.1 AXCM01001720 AAAB01008846 EAA06228.5 APCN01004026 ADMH02001970 ETN60126.1 ATLV01013426 KE524855 KFB37965.1 AJWK01006356 AXCN02001227 UFQT01000222 SSX21944.1 UFQS01000634 UFQT01000634 SSX05607.1 SSX25966.1 DS235053 EEB11083.1 EDS38294.1 UFQS01000368 UFQT01000368 SSX03231.1 SSX23597.1 GEDC01029520 JAS07778.1 EAT43887.1 KK852782 KDR16611.1 JXUM01031748 JXUM01031749 KQ560934 KXJ80286.1 PYGN01000367 PSN47583.1 GAKP01007983 GAKP01007981 JAC50969.1 GDHF01031181 GDHF01019229 GDHF01009047 JAI21133.1 JAI33085.1 JAI43267.1 OUUW01000006 SPP82205.1 CH379061 KRT04640.1 NEVH01007828 PNF35079.1 SPP82209.1 SPP82208.1 GDHC01010417 JAQ08212.1 GDHF01014567 JAI37747.1 KK854651 PTY17646.1 SPP82207.1 SPP82206.1 GL450828 EFN80313.1 CH479189 EDW25311.1 AE014134 BT003187 AAN10385.1 AAO24942.1 KK107453 QOIP01000004 EZA50595.1 RLU24169.1 EAL32988.3 GBXI01008607 JAD05685.1 JR039392 JR039393 AEY58671.1 AEY58672.1 AHN54096.1 CM000157 KRJ97183.1 CH954177 KQS70032.1 JRES01001163 KNC24937.1 GBXI01001230 JAD13062.1 CM002910 KMY87836.1 GL888932 EGI57319.1 GEDC01021123 JAS16175.1 KMY87837.1 SPP82210.1 CCAG010022353 KQ434778 KZC04336.1 NNAY01002084 OXU22132.1 CH902624 KPU74206.1 CH964239 EDW82120.2 KQ414638 KOC66914.1 BABH01001086 NWSH01001609 PCG70709.1 AHN54095.1 AY058246 AAF51132.3 AAL13475.1 GAMC01005517 JAC01039.1 JXJN01006969
GDQN01010297 GDQN01004622 JAT80757.1 JAT86432.1 AGBW02013857 OWR42629.1 RSAL01000413 RVE41839.1 JTDY01005896 KOB66504.1 GALX01003816 JAB64650.1 KQ971338 EFA01890.1 APGK01010729 KB738283 ENN82679.1 AJVK01018277 AJVK01018278 GEZM01054482 JAV73621.1 KB632278 ERL91229.1 GFDF01011173 JAV02911.1 GFDF01011191 JAV02893.1 CH477312 EAT43886.1 GFDF01011172 JAV02912.1 DS232243 EDS38295.1 GDAI01001110 JAI16493.1 CVRI01000035 CRK92831.1 AXCM01001720 AAAB01008846 EAA06228.5 APCN01004026 ADMH02001970 ETN60126.1 ATLV01013426 KE524855 KFB37965.1 AJWK01006356 AXCN02001227 UFQT01000222 SSX21944.1 UFQS01000634 UFQT01000634 SSX05607.1 SSX25966.1 DS235053 EEB11083.1 EDS38294.1 UFQS01000368 UFQT01000368 SSX03231.1 SSX23597.1 GEDC01029520 JAS07778.1 EAT43887.1 KK852782 KDR16611.1 JXUM01031748 JXUM01031749 KQ560934 KXJ80286.1 PYGN01000367 PSN47583.1 GAKP01007983 GAKP01007981 JAC50969.1 GDHF01031181 GDHF01019229 GDHF01009047 JAI21133.1 JAI33085.1 JAI43267.1 OUUW01000006 SPP82205.1 CH379061 KRT04640.1 NEVH01007828 PNF35079.1 SPP82209.1 SPP82208.1 GDHC01010417 JAQ08212.1 GDHF01014567 JAI37747.1 KK854651 PTY17646.1 SPP82207.1 SPP82206.1 GL450828 EFN80313.1 CH479189 EDW25311.1 AE014134 BT003187 AAN10385.1 AAO24942.1 KK107453 QOIP01000004 EZA50595.1 RLU24169.1 EAL32988.3 GBXI01008607 JAD05685.1 JR039392 JR039393 AEY58671.1 AEY58672.1 AHN54096.1 CM000157 KRJ97183.1 CH954177 KQS70032.1 JRES01001163 KNC24937.1 GBXI01001230 JAD13062.1 CM002910 KMY87836.1 GL888932 EGI57319.1 GEDC01021123 JAS16175.1 KMY87837.1 SPP82210.1 CCAG010022353 KQ434778 KZC04336.1 NNAY01002084 OXU22132.1 CH902624 KPU74206.1 CH964239 EDW82120.2 KQ414638 KOC66914.1 BABH01001086 NWSH01001609 PCG70709.1 AHN54095.1 AY058246 AAF51132.3 AAL13475.1 GAMC01005517 JAC01039.1 JXJN01006969
Proteomes
UP000053240
UP000053268
UP000007151
UP000283053
UP000037510
UP000007266
+ More
UP000019118 UP000092462 UP000030742 UP000008820 UP000002320 UP000075885 UP000183832 UP000075883 UP000007062 UP000076407 UP000075840 UP000075880 UP000000673 UP000069272 UP000030765 UP000092461 UP000076408 UP000075886 UP000075900 UP000075902 UP000075920 UP000075884 UP000009046 UP000027135 UP000069940 UP000249989 UP000245037 UP000268350 UP000095300 UP000001819 UP000235965 UP000095301 UP000005203 UP000192221 UP000008237 UP000008744 UP000091820 UP000000803 UP000053097 UP000279307 UP000002282 UP000008711 UP000037069 UP000007755 UP000092444 UP000076502 UP000215335 UP000007801 UP000007798 UP000053825 UP000005204 UP000218220 UP000092443 UP000092460 UP000078200
UP000019118 UP000092462 UP000030742 UP000008820 UP000002320 UP000075885 UP000183832 UP000075883 UP000007062 UP000076407 UP000075840 UP000075880 UP000000673 UP000069272 UP000030765 UP000092461 UP000076408 UP000075886 UP000075900 UP000075902 UP000075920 UP000075884 UP000009046 UP000027135 UP000069940 UP000249989 UP000245037 UP000268350 UP000095300 UP000001819 UP000235965 UP000095301 UP000005203 UP000192221 UP000008237 UP000008744 UP000091820 UP000000803 UP000053097 UP000279307 UP000002282 UP000008711 UP000037069 UP000007755 UP000092444 UP000076502 UP000215335 UP000007801 UP000007798 UP000053825 UP000005204 UP000218220 UP000092443 UP000092460 UP000078200
Interpro
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
A0A2H1VCH1
A0A194RHF9
A0A194PJI1
A0A1E1W187
A0A212EMC9
A0A3S2KYZ8
+ More
A0A0L7KTX5 V5G3P9 D2A3K9 N6TQA5 A0A1B0DPH1 A0A1Y1LL30 U4UBC1 A0A1L8D8X6 A0A1L8D8V6 Q0IFL7 A0A1L8D8X9 B0X0Q3 A0A182PKX5 A0A0K8TQR5 A0A1J1HXL1 A0A182MBX3 Q7QEN7 A0A182XES1 A0A182HIN8 A0A182JKA1 W5J828 A0A182FJD8 A0A084VJ21 A0A1B0ETG4 A0A182YCN0 A0A182Q0E2 A0A182RZ31 A0A182U4E0 A0A336LVG2 A0A336KML1 A0A182VVP7 A0A182N028 E0VCH7 B0X0Q2 A0A336M5E8 A0A1B6C2N4 A0A1S4F8C6 Q0IFL6 A0A067R125 A0A182HB61 A0A2P8YTK7 A0A034W8D4 A0A0K8V3B4 A0A3B0JIZ8 A0A1I8NPZ8 A0A0R3P033 A0A2J7R2L2 A0A3B0K850 A0A3B0KF18 A0A146LN45 A0A1I8M6Y2 A0A1I8M6Y3 A0A088AUK5 A0A0K8VFW9 A0A1W4V6L7 A0A1W4UU99 A0A2R7WDC1 A0A3B0JIL3 A0A3B0K9A0 E2BVD7 B4GSF2 A0A1A9WCG6 Q8IQ02 A0A026W3F3 Q29L36 A0A0A1X4K0 V9IDZ6 X2J8I0 A0A0R1DJ26 A0A0Q5W8X6 A0A0L0BY36 A0A1W4UUN8 A0A0A1XQ32 A0A0J9QUY9 F4X8Q0 A0A1B6CS77 A0A0J9QUT9 A0A3B0K2U2 A0A1B0FRD1 A0A154NXD9 A0A232EUU7 A0A0P9A708 B4NEF1 A0A0L7R7U6 H9IVH9 A0A2A4JFV7 X2JD17 Q9VQN3 A0A1A9YQR1 W8BVT8 A0A1B0B143 A0A1A9UZS0
A0A0L7KTX5 V5G3P9 D2A3K9 N6TQA5 A0A1B0DPH1 A0A1Y1LL30 U4UBC1 A0A1L8D8X6 A0A1L8D8V6 Q0IFL7 A0A1L8D8X9 B0X0Q3 A0A182PKX5 A0A0K8TQR5 A0A1J1HXL1 A0A182MBX3 Q7QEN7 A0A182XES1 A0A182HIN8 A0A182JKA1 W5J828 A0A182FJD8 A0A084VJ21 A0A1B0ETG4 A0A182YCN0 A0A182Q0E2 A0A182RZ31 A0A182U4E0 A0A336LVG2 A0A336KML1 A0A182VVP7 A0A182N028 E0VCH7 B0X0Q2 A0A336M5E8 A0A1B6C2N4 A0A1S4F8C6 Q0IFL6 A0A067R125 A0A182HB61 A0A2P8YTK7 A0A034W8D4 A0A0K8V3B4 A0A3B0JIZ8 A0A1I8NPZ8 A0A0R3P033 A0A2J7R2L2 A0A3B0K850 A0A3B0KF18 A0A146LN45 A0A1I8M6Y2 A0A1I8M6Y3 A0A088AUK5 A0A0K8VFW9 A0A1W4V6L7 A0A1W4UU99 A0A2R7WDC1 A0A3B0JIL3 A0A3B0K9A0 E2BVD7 B4GSF2 A0A1A9WCG6 Q8IQ02 A0A026W3F3 Q29L36 A0A0A1X4K0 V9IDZ6 X2J8I0 A0A0R1DJ26 A0A0Q5W8X6 A0A0L0BY36 A0A1W4UUN8 A0A0A1XQ32 A0A0J9QUY9 F4X8Q0 A0A1B6CS77 A0A0J9QUT9 A0A3B0K2U2 A0A1B0FRD1 A0A154NXD9 A0A232EUU7 A0A0P9A708 B4NEF1 A0A0L7R7U6 H9IVH9 A0A2A4JFV7 X2JD17 Q9VQN3 A0A1A9YQR1 W8BVT8 A0A1B0B143 A0A1A9UZS0
PDB
6O84
E-value=4.30958e-10,
Score=157
Ontologies
PANTHER
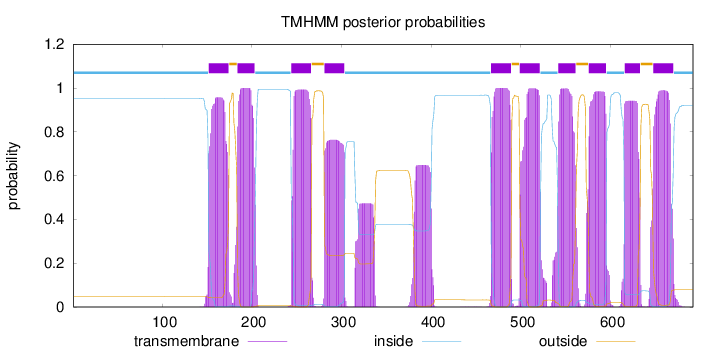
Topology
Length:
692
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
230.03817
Exp number, first 60 AAs:
0
Total prob of N-in:
0.95316
inside
1 - 151
TMhelix
152 - 174
outside
175 - 183
TMhelix
184 - 203
inside
204 - 243
TMhelix
244 - 266
outside
267 - 280
TMhelix
281 - 303
inside
304 - 466
TMhelix
467 - 489
outside
490 - 498
TMhelix
499 - 521
inside
522 - 541
TMhelix
542 - 561
outside
562 - 575
TMhelix
576 - 595
inside
596 - 615
TMhelix
616 - 633
outside
634 - 647
TMhelix
648 - 670
inside
671 - 692
Population Genetic Test Statistics
Pi
20.477827
Theta
164.925582
Tajima's D
0.431889
CLR
0.727903
CSRT
0.496675166241688
Interpretation
Uncertain
