Gene
KWMTBOMO07793
Annotation
PREDICTED:_uncharacterized_protein_LOC101737731_[Bombyx_mori]
Transcription factor
Location in the cell
Extracellular Reliability : 1.332 Mitochondrial Reliability : 1.64 Nuclear Reliability : 1.151
Sequence
CDS
ATGCCAGCATGTGTAGTGAAGTGGTGCAGGAGCCATTCAAGGTTAAAATGCCCAGGGATCACTTTTCACTATTTTCCTGCTGATGCAATTCGGCAAAAAAAGTGGATAGCTGCAGTGAAGTTGGAAAGACAGGAATATGACTGGATGCCTGTAAAAACTAGCAGAGTATGCTCTATTCATTTTGATAAAAAAGATTTTTACAAATCATTAAAAGGTTTATACAATGTTACACAAAACTGCAGTAACAATTTGTACAATGATTCGTTTTGA
Protein
MPACVVKWCRSHSRLKCPGITFHYFPADAIRQKKWIAAVKLERQEYDWMPVKTSRVCSIHFDKKDFYKSLKGLYNVTQNCSNNLYNDSF
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A3S2PH61
A0A3S2KYQ6
A0A3S2P1T5
A0A2H1WLY2
A0A3S2LJC3
A0A3S2NMW6
+ More
A0A3S2LR03 A0A2A4K4M7 A0A3S2P0Q1 A0A0K8RG12 A0A3S2L6Q1 A0A3S2NTY9 A0A194Q3V2 A0A0L7KTK8 A0A131XNN7 A0A2P8YK78 A0A3S2LM39 H9JHG4 A0A2H1WUK2 A0A3S2LWX1 A0A1E1W0L8 A0A1E1W6J1 A0A1B6JLM8 A0A224YYJ7 A0A194R6F3 A0A131YI94 A0A1E1VYI8 A0A224Z0Z7 A0A3S2L852 A0A224YVD2 A0A224Z0U4 A0A2P8XN38 A0A2P8XN62 T1IW91 A0A0A9Y1L2 A0A1Z5L7W1 A0A3S2TIX3 A0A3S2LSN2 A0A2H1WZ56 A0A194QBV1 A0A3S2LE61 A0A293LGB5 A0A2H1VW64 A0A1E1XNZ8 A0A194Q151 A0A131YL35 A0A147BC10 A0A131YST0 A0A087TW15 A0A224Z0Y6 A0A1E1XNW3 A0A087UU52 A0A2S2NEE6 A0A3Q3EX00 A0A131XP92 A0A194R2I1 V5H8E7 A0A3S2L9A0 A0A224Z483 A0A0N1PIG4 A0A3S2M4I9 A0A293MHT8 A0A1B0CBA5 A0A1B6DGS4 A0A1L8DIP4 A0A3S2LLA7 A0A194QVL0 A0A293LL46 A0A146LEM6 F4WUC5 C1BNR4 A0A026X1G5 A0A3L8DTM9 A0A194Q527 A0A1E1VXJ3 A0A1E1WQV1 A0A158NQF8 A0A3B3I0M3 A0A3B3HDE6 A0A147BG09 A0A3B3BY40 A0A1Z5L812 A0A2H1VHN5 A0A3B1J269 A0A147BB70 V5HVR1
A0A3S2LR03 A0A2A4K4M7 A0A3S2P0Q1 A0A0K8RG12 A0A3S2L6Q1 A0A3S2NTY9 A0A194Q3V2 A0A0L7KTK8 A0A131XNN7 A0A2P8YK78 A0A3S2LM39 H9JHG4 A0A2H1WUK2 A0A3S2LWX1 A0A1E1W0L8 A0A1E1W6J1 A0A1B6JLM8 A0A224YYJ7 A0A194R6F3 A0A131YI94 A0A1E1VYI8 A0A224Z0Z7 A0A3S2L852 A0A224YVD2 A0A224Z0U4 A0A2P8XN38 A0A2P8XN62 T1IW91 A0A0A9Y1L2 A0A1Z5L7W1 A0A3S2TIX3 A0A3S2LSN2 A0A2H1WZ56 A0A194QBV1 A0A3S2LE61 A0A293LGB5 A0A2H1VW64 A0A1E1XNZ8 A0A194Q151 A0A131YL35 A0A147BC10 A0A131YST0 A0A087TW15 A0A224Z0Y6 A0A1E1XNW3 A0A087UU52 A0A2S2NEE6 A0A3Q3EX00 A0A131XP92 A0A194R2I1 V5H8E7 A0A3S2L9A0 A0A224Z483 A0A0N1PIG4 A0A3S2M4I9 A0A293MHT8 A0A1B0CBA5 A0A1B6DGS4 A0A1L8DIP4 A0A3S2LLA7 A0A194QVL0 A0A293LL46 A0A146LEM6 F4WUC5 C1BNR4 A0A026X1G5 A0A3L8DTM9 A0A194Q527 A0A1E1VXJ3 A0A1E1WQV1 A0A158NQF8 A0A3B3I0M3 A0A3B3HDE6 A0A147BG09 A0A3B3BY40 A0A1Z5L812 A0A2H1VHN5 A0A3B1J269 A0A147BB70 V5HVR1
Pubmed
EMBL
RSAL01000035
RVE51327.1
RSAL01000469
RVE41615.1
RSAL01000002
RVE54928.1
+ More
ODYU01009475 SOQ53942.1 RSAL01000001 RVE55133.1 RSAL01004978 RVE40066.1 RSAL01000381 RVE42052.1 NWSH01000136 PCG79197.1 RSAL01000065 RVE49415.1 GADI01003716 JAA70092.1 RSAL01003087 RVE40344.1 RSAL01000082 RVE48523.1 KQ459586 KPI98085.1 JTDY01006020 KOB66364.1 GEFM01006752 JAP69044.1 PYGN01000534 PSN44655.1 RSAL01000060 RVE49645.1 BABH01012529 ODYU01011169 SOQ56745.1 RSAL01000140 RVE46105.1 GDQN01010514 JAT80540.1 GDQN01008480 JAT82574.1 GECU01007568 JAT00139.1 GFPF01008545 MAA19691.1 KQ460685 KPJ12820.1 GEDV01010355 JAP78202.1 GDQN01011302 JAT79752.1 GFPF01008544 MAA19690.1 RSAL01000093 RVE47898.1 GFPF01008543 MAA19689.1 GFPF01008546 MAA19692.1 PYGN01001677 PSN33423.1 PSN33425.1 JH431611 GBHO01017555 JAG26049.1 GFJQ02003454 JAW03516.1 RSAL01000106 RVE47304.1 RSAL01000016 RVE53005.1 ODYU01012114 SOQ58236.1 KQ459232 KPJ02470.1 RSAL01000187 RVE44781.1 GFWV01003053 MAA27783.1 ODYU01004808 SOQ45070.1 GFAA01002387 JAU01048.1 KQ459580 KPI99281.1 GEDV01008538 JAP80019.1 GEGO01007106 JAR88298.1 GEDV01007391 JAP81166.1 KK117010 KFM69304.1 GFPF01008534 MAA19680.1 GFAA01002414 JAU01021.1 KK121633 KFM80891.1 GGMR01002982 MBY15601.1 GEFM01006753 JAP69043.1 KQ460845 KPJ11998.1 GANP01005028 JAB79440.1 RSAL01000412 RVE41849.1 GFPF01012839 MAA23985.1 KQ460396 KPJ15350.1 RSAL01000037 RVE51214.1 GFWV01015725 MAA40454.1 AJWK01004933 GEDC01012411 JAS24887.1 GFDF01007758 JAV06326.1 RSAL01011042 RVE39731.1 KQ461181 KPJ07591.1 GFWV01009498 MAA34227.1 GDHC01013187 JAQ05442.1 GL888355 EGI62233.1 BT076243 ACO10667.1 KK107063 EZA61229.1 QOIP01000005 RLU23118.1 KQ459463 KPJ00464.1 GDQN01011663 JAT79391.1 GDQN01001641 JAT89413.1 ADTU01023218 ADTU01023219 GEGO01005730 JAR89674.1 GFJQ02003642 JAW03328.1 ODYU01002370 SOQ39784.1 GEGO01007413 JAR87991.1 GANP01001799 JAB82669.1
ODYU01009475 SOQ53942.1 RSAL01000001 RVE55133.1 RSAL01004978 RVE40066.1 RSAL01000381 RVE42052.1 NWSH01000136 PCG79197.1 RSAL01000065 RVE49415.1 GADI01003716 JAA70092.1 RSAL01003087 RVE40344.1 RSAL01000082 RVE48523.1 KQ459586 KPI98085.1 JTDY01006020 KOB66364.1 GEFM01006752 JAP69044.1 PYGN01000534 PSN44655.1 RSAL01000060 RVE49645.1 BABH01012529 ODYU01011169 SOQ56745.1 RSAL01000140 RVE46105.1 GDQN01010514 JAT80540.1 GDQN01008480 JAT82574.1 GECU01007568 JAT00139.1 GFPF01008545 MAA19691.1 KQ460685 KPJ12820.1 GEDV01010355 JAP78202.1 GDQN01011302 JAT79752.1 GFPF01008544 MAA19690.1 RSAL01000093 RVE47898.1 GFPF01008543 MAA19689.1 GFPF01008546 MAA19692.1 PYGN01001677 PSN33423.1 PSN33425.1 JH431611 GBHO01017555 JAG26049.1 GFJQ02003454 JAW03516.1 RSAL01000106 RVE47304.1 RSAL01000016 RVE53005.1 ODYU01012114 SOQ58236.1 KQ459232 KPJ02470.1 RSAL01000187 RVE44781.1 GFWV01003053 MAA27783.1 ODYU01004808 SOQ45070.1 GFAA01002387 JAU01048.1 KQ459580 KPI99281.1 GEDV01008538 JAP80019.1 GEGO01007106 JAR88298.1 GEDV01007391 JAP81166.1 KK117010 KFM69304.1 GFPF01008534 MAA19680.1 GFAA01002414 JAU01021.1 KK121633 KFM80891.1 GGMR01002982 MBY15601.1 GEFM01006753 JAP69043.1 KQ460845 KPJ11998.1 GANP01005028 JAB79440.1 RSAL01000412 RVE41849.1 GFPF01012839 MAA23985.1 KQ460396 KPJ15350.1 RSAL01000037 RVE51214.1 GFWV01015725 MAA40454.1 AJWK01004933 GEDC01012411 JAS24887.1 GFDF01007758 JAV06326.1 RSAL01011042 RVE39731.1 KQ461181 KPJ07591.1 GFWV01009498 MAA34227.1 GDHC01013187 JAQ05442.1 GL888355 EGI62233.1 BT076243 ACO10667.1 KK107063 EZA61229.1 QOIP01000005 RLU23118.1 KQ459463 KPJ00464.1 GDQN01011663 JAT79391.1 GDQN01001641 JAT89413.1 ADTU01023218 ADTU01023219 GEGO01005730 JAR89674.1 GFJQ02003642 JAW03328.1 ODYU01002370 SOQ39784.1 GEGO01007413 JAR87991.1 GANP01001799 JAB82669.1
Proteomes
PRIDE
Pfam
Interpro
IPR038441
THAP_Znf_sf
+ More
IPR006612 THAP_Znf
IPR021896 Transposase_37
IPR026516 THAP1
IPR027124 Swc5/CFDP2
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011009 Kinase-like_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR031961 DUF4780
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
IPR006612 THAP_Znf
IPR021896 Transposase_37
IPR026516 THAP1
IPR027124 Swc5/CFDP2
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR011009 Kinase-like_dom_sf
IPR020635 Tyr_kinase_cat_dom
IPR008266 Tyr_kinase_AS
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR000719 Prot_kinase_dom
IPR017441 Protein_kinase_ATP_BS
IPR031961 DUF4780
IPR036236 Znf_C2H2_sf
IPR013087 Znf_C2H2_type
IPR012934 Znf_AD
Gene 3D
ProteinModelPortal
A0A3S2PH61
A0A3S2KYQ6
A0A3S2P1T5
A0A2H1WLY2
A0A3S2LJC3
A0A3S2NMW6
+ More
A0A3S2LR03 A0A2A4K4M7 A0A3S2P0Q1 A0A0K8RG12 A0A3S2L6Q1 A0A3S2NTY9 A0A194Q3V2 A0A0L7KTK8 A0A131XNN7 A0A2P8YK78 A0A3S2LM39 H9JHG4 A0A2H1WUK2 A0A3S2LWX1 A0A1E1W0L8 A0A1E1W6J1 A0A1B6JLM8 A0A224YYJ7 A0A194R6F3 A0A131YI94 A0A1E1VYI8 A0A224Z0Z7 A0A3S2L852 A0A224YVD2 A0A224Z0U4 A0A2P8XN38 A0A2P8XN62 T1IW91 A0A0A9Y1L2 A0A1Z5L7W1 A0A3S2TIX3 A0A3S2LSN2 A0A2H1WZ56 A0A194QBV1 A0A3S2LE61 A0A293LGB5 A0A2H1VW64 A0A1E1XNZ8 A0A194Q151 A0A131YL35 A0A147BC10 A0A131YST0 A0A087TW15 A0A224Z0Y6 A0A1E1XNW3 A0A087UU52 A0A2S2NEE6 A0A3Q3EX00 A0A131XP92 A0A194R2I1 V5H8E7 A0A3S2L9A0 A0A224Z483 A0A0N1PIG4 A0A3S2M4I9 A0A293MHT8 A0A1B0CBA5 A0A1B6DGS4 A0A1L8DIP4 A0A3S2LLA7 A0A194QVL0 A0A293LL46 A0A146LEM6 F4WUC5 C1BNR4 A0A026X1G5 A0A3L8DTM9 A0A194Q527 A0A1E1VXJ3 A0A1E1WQV1 A0A158NQF8 A0A3B3I0M3 A0A3B3HDE6 A0A147BG09 A0A3B3BY40 A0A1Z5L812 A0A2H1VHN5 A0A3B1J269 A0A147BB70 V5HVR1
A0A3S2LR03 A0A2A4K4M7 A0A3S2P0Q1 A0A0K8RG12 A0A3S2L6Q1 A0A3S2NTY9 A0A194Q3V2 A0A0L7KTK8 A0A131XNN7 A0A2P8YK78 A0A3S2LM39 H9JHG4 A0A2H1WUK2 A0A3S2LWX1 A0A1E1W0L8 A0A1E1W6J1 A0A1B6JLM8 A0A224YYJ7 A0A194R6F3 A0A131YI94 A0A1E1VYI8 A0A224Z0Z7 A0A3S2L852 A0A224YVD2 A0A224Z0U4 A0A2P8XN38 A0A2P8XN62 T1IW91 A0A0A9Y1L2 A0A1Z5L7W1 A0A3S2TIX3 A0A3S2LSN2 A0A2H1WZ56 A0A194QBV1 A0A3S2LE61 A0A293LGB5 A0A2H1VW64 A0A1E1XNZ8 A0A194Q151 A0A131YL35 A0A147BC10 A0A131YST0 A0A087TW15 A0A224Z0Y6 A0A1E1XNW3 A0A087UU52 A0A2S2NEE6 A0A3Q3EX00 A0A131XP92 A0A194R2I1 V5H8E7 A0A3S2L9A0 A0A224Z483 A0A0N1PIG4 A0A3S2M4I9 A0A293MHT8 A0A1B0CBA5 A0A1B6DGS4 A0A1L8DIP4 A0A3S2LLA7 A0A194QVL0 A0A293LL46 A0A146LEM6 F4WUC5 C1BNR4 A0A026X1G5 A0A3L8DTM9 A0A194Q527 A0A1E1VXJ3 A0A1E1WQV1 A0A158NQF8 A0A3B3I0M3 A0A3B3HDE6 A0A147BG09 A0A3B3BY40 A0A1Z5L812 A0A2H1VHN5 A0A3B1J269 A0A147BB70 V5HVR1
PDB
2D8R
E-value=0.0263801,
Score=81
Ontologies
KEGG
GO
PANTHER
Topology
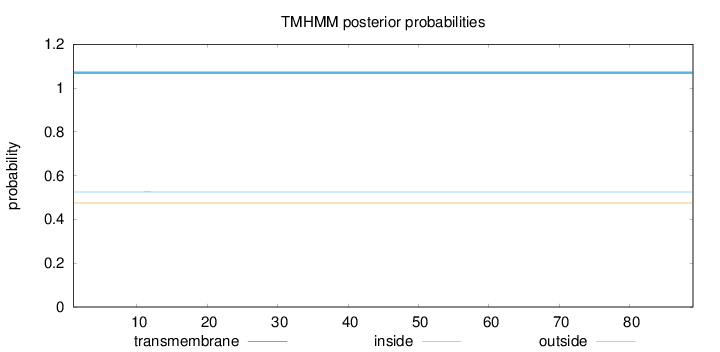
Length:
89
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00129
Exp number, first 60 AAs:
0.00129
Total prob of N-in:
0.52556
inside
1 - 89
Population Genetic Test Statistics
Pi
131.797
Theta
139.242946
Tajima's D
-0.14992
CLR
3.069783
CSRT
0.327083645817709
Interpretation
Uncertain
