Pre Gene Modal
BGIBMGA001236
Annotation
PREDICTED:_succinate_dehydrogenase_assembly_factor_2-B?_mitochondrial-like_[Plutella_xylostella]
Full name
Succinate dehydrogenase assembly factor 2, mitochondrial
+ More
Succinate dehydrogenase assembly factor 2-B, mitochondrial
Succinate dehydrogenase assembly factor 2-A, mitochondrial
Succinate dehydrogenase assembly factor 2-B, mitochondrial
Succinate dehydrogenase assembly factor 2-A, mitochondrial
Location in the cell
Mitochondrial Reliability : 1.954 Nuclear Reliability : 1.887
Sequence
CDS
ATGTTGCATACTCGAACCCTTAACCTTTCTTATAATGCACTCAAAAATGTCCAGAAAGTGGCTTTTTCAAGTAATACCACCAACAACATGGATACAACCTACATGGAGATTCCTCTTTATGATGTTGAGAAACAACAACCGCTGGAGAAACGTAAAGCAAGATTGCATTACCAATCGCGAAAAAGAGGAATGTTAGAAAACGATCTTCTACTCAGCACTTTTGCGAAGAAGTATTTGAATGGTTTTAATGAAAAACAGACTATGATGTACGATCGTCTCATAAACTCACCCAGCAACGATTGGGACATATTCTATTGGATAGTTGAAAAGAAGCCAACTCCTAAAGAATTTGACAATGAAATTATGAATCTTCTCAAAAAACATGCTAAAAATGAAGATAAAATTGCACTCCGACAACCCGATCTACATTAA
Protein
MLHTRTLNLSYNALKNVQKVAFSSNTTNNMDTTYMEIPLYDVEKQQPLEKRKARLHYQSRKRGMLENDLLLSTFAKKYLNGFNEKQTMMYDRLINSPSNDWDIFYWIVEKKPTPKEFDNEIMNLLKKHAKNEDKIALRQPDLH
Summary
Description
Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol. Required for flavinylation (covalent attachment of FAD) of the flavoprotein subunit of the SDH catalytic dimer.
Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol. Required for flavinylation (covalent attachment of FAD) of the flavoprotein subunit SDHA of the SDH catalytic dimer.
Plays an essential role in the assembly of succinate dehydrogenase (SDH), an enzyme complex (also referred to as respiratory complex II) that is a component of both the tricarboxylic acid (TCA) cycle and the mitochondrial electron transport chain, and which couples the oxidation of succinate to fumarate with the reduction of ubiquinone (coenzyme Q) to ubiquinol. Required for flavinylation (covalent attachment of FAD) of the flavoprotein subunit SDHA of the SDH catalytic dimer.
Subunit
Interacts with the flavoprotein subunit within the SDH catalytic dimer.
Interacts with SDHA within the SDH catalytic dimer.
Interacts with SDHA within the SDH catalytic dimer.
Miscellaneous
This protein may be expected to contain an N-terminal transit peptide but none has been predicted.
Similarity
Belongs to the SDHAF2 family.
Keywords
Chaperone
Complete proteome
Mitochondrion
Reference proteome
Transit peptide
Feature
chain Succinate dehydrogenase assembly factor 2-B, mitochondrial
Uniprot
H9IVF6
I4DPA5
A0A194RGZ9
A0A212FI19
A0A1E1WBP0
A0A2A4JSL7
+ More
A0A2H1VZZ1 B4N665 A0A0M4EHY4 E0VKE1 B3MGU5 A0A0J9TW46 B4QFP7 B4HRL4 A0A1B2AJ40 Q4V5I9 A0A0M3QUL9 A0A1B0DEP3 A0A1W4VPE7 B4P2P8 B4GDB3 B5DZ31 A0A3B0J0G1 B3MI37 A0A2M4ARD1 A0A2M4AMC5 B3N8S9 B4NXN5 A0A0B4KF96 A1Z897 A0A2M4ARF0 B4HMQ1 A0A2M3ZIP8 B5E0U2 A0A0J9R9P0 A0A3B0JM63 H0RNF9 B4QID8 A0A0L7RFV2 K7IQX4 A0A1W4WAK4 B3N6D9 A0A232EUM4 B4GG58 T1EA61 A0A182N346 A0A182QH38 W5JQ07 A0A2M4C275 A0A1B0CSA2 B4LKE5 A0A154PBQ2 A0A182FVN5 D6WAG2 A0A232EYZ0 V9IM23 A0A2A3EC49 C3ZTP0 C3Y5H2 K7J567 A0A182RW68 A0A182YMI9 A0A0J7L6U8 A0A026W3T6 A0A0Q9WCX0 A0A182TCJ5 A0A3M6U3M1 K1S2Y6 A0A182PGE3 A0A158NW55 B4KN44 Q7Q014 A0A182TXH6 A0A182HPU6 A0A3S3P7A0 A0A182WZY2 A0A084WUM3 A0A067QGL4 B4J7I7 A0A182W2A0 A0A182K9U5 B4MRE7 A0A182UN54 A0A182KSX9 B0XK69 A0A023EHW8 A0A151WVC4 A0A195B8A9 F4WNF1 A0A1Q3FMC0 A0A1L8E027 A0A0K8TQI4 H3ABN6 A0A182GHL8 A0A151IM66 A0A2B4SJ61 A0A2K7P6A1
A0A2H1VZZ1 B4N665 A0A0M4EHY4 E0VKE1 B3MGU5 A0A0J9TW46 B4QFP7 B4HRL4 A0A1B2AJ40 Q4V5I9 A0A0M3QUL9 A0A1B0DEP3 A0A1W4VPE7 B4P2P8 B4GDB3 B5DZ31 A0A3B0J0G1 B3MI37 A0A2M4ARD1 A0A2M4AMC5 B3N8S9 B4NXN5 A0A0B4KF96 A1Z897 A0A2M4ARF0 B4HMQ1 A0A2M3ZIP8 B5E0U2 A0A0J9R9P0 A0A3B0JM63 H0RNF9 B4QID8 A0A0L7RFV2 K7IQX4 A0A1W4WAK4 B3N6D9 A0A232EUM4 B4GG58 T1EA61 A0A182N346 A0A182QH38 W5JQ07 A0A2M4C275 A0A1B0CSA2 B4LKE5 A0A154PBQ2 A0A182FVN5 D6WAG2 A0A232EYZ0 V9IM23 A0A2A3EC49 C3ZTP0 C3Y5H2 K7J567 A0A182RW68 A0A182YMI9 A0A0J7L6U8 A0A026W3T6 A0A0Q9WCX0 A0A182TCJ5 A0A3M6U3M1 K1S2Y6 A0A182PGE3 A0A158NW55 B4KN44 Q7Q014 A0A182TXH6 A0A182HPU6 A0A3S3P7A0 A0A182WZY2 A0A084WUM3 A0A067QGL4 B4J7I7 A0A182W2A0 A0A182K9U5 B4MRE7 A0A182UN54 A0A182KSX9 B0XK69 A0A023EHW8 A0A151WVC4 A0A195B8A9 F4WNF1 A0A1Q3FMC0 A0A1L8E027 A0A0K8TQI4 H3ABN6 A0A182GHL8 A0A151IM66 A0A2B4SJ61 A0A2K7P6A1
Pubmed
19121390
22651552
26354079
22118469
17994087
20566863
+ More
22936249 10731132 12537572 15632085 12537568 12537573 12537574 16110336 17569856 17569867 12537569 20075255 28648823 20920257 23761445 18362917 19820115 18563158 25244985 24508170 30382153 22992520 21347285 12364791 24438588 24845553 20966253 24945155 21719571 26369729 9215903 26483478
22936249 10731132 12537572 15632085 12537568 12537573 12537574 16110336 17569856 17569867 12537569 20075255 28648823 20920257 23761445 18362917 19820115 18563158 25244985 24508170 30382153 22992520 21347285 12364791 24438588 24845553 20966253 24945155 21719571 26369729 9215903 26483478
EMBL
BABH01001025
AK403496
KQ459606
BAM19745.1
KPI91231.1
KQ460205
+ More
KPJ16862.1 AGBW02008449 OWR53378.1 GDQN01006695 JAT84359.1 NWSH01000724 PCG74604.1 ODYU01005495 SOQ46405.1 CH964154 CP012524 ALC40605.1 DS235243 EEB13857.1 CH902619 CM002911 KMY92225.1 CM000362 CH480816 KX531364 ANY27174.1 AE013599 BT022667 ALC40899.1 AJVK01014883 CM000157 CH479181 CM000071 OUUW01000001 SPP74107.1 GGFK01010034 MBW43355.1 GGFK01008608 MBW41929.1 CH954177 AGB93401.1 AY071255 GGFK01010054 MBW43375.1 GGFM01007567 MBW28318.1 KMY92788.1 SPP73731.1 BT132881 AEV23905.1 KQ414606 KOC69611.1 NNAY01002107 OXU22059.1 CH479183 GAMD01000717 JAB00874.1 AXCN02001125 ADMH02000489 ETN66221.1 GGFJ01010233 MBW59374.1 AJWK01025830 CH940648 KQ434869 KZC09237.1 KQ971312 EEZ98628.2 NNAY01001588 OXU23510.1 JR051745 AEY61551.1 KZ288293 PBC29054.1 GG666679 EEN44038.1 GG666487 EEN64219.1 AAZX01005033 LBMM01000423 KMQ98396.1 KK107447 EZA50750.1 KRF79633.1 RCHS01002309 RMX48134.1 JH816866 EKC41731.1 ADTU01027818 CH933808 AAAB01008986 EAA00134.4 APCN01003272 NCKU01000233 RWS16423.1 ATLV01027100 KE525423 KFB53917.1 KK853502 KDR07129.1 CH916367 EDW01111.1 CH963850 DS233727 GAPW01005159 JAC08439.1 KQ982706 KYQ51823.1 KQ976565 KYM80434.1 GL888237 EGI64144.1 GFDL01006285 JAV28760.1 GFDF01002050 JAV12034.1 GDAI01000976 JAI16627.1 AFYH01010809 JXUM01063949 JXUM01063950 KQ562275 KXJ76259.1 KQ977063 KYN05977.1 LSMT01000080 PFX28608.1
KPJ16862.1 AGBW02008449 OWR53378.1 GDQN01006695 JAT84359.1 NWSH01000724 PCG74604.1 ODYU01005495 SOQ46405.1 CH964154 CP012524 ALC40605.1 DS235243 EEB13857.1 CH902619 CM002911 KMY92225.1 CM000362 CH480816 KX531364 ANY27174.1 AE013599 BT022667 ALC40899.1 AJVK01014883 CM000157 CH479181 CM000071 OUUW01000001 SPP74107.1 GGFK01010034 MBW43355.1 GGFK01008608 MBW41929.1 CH954177 AGB93401.1 AY071255 GGFK01010054 MBW43375.1 GGFM01007567 MBW28318.1 KMY92788.1 SPP73731.1 BT132881 AEV23905.1 KQ414606 KOC69611.1 NNAY01002107 OXU22059.1 CH479183 GAMD01000717 JAB00874.1 AXCN02001125 ADMH02000489 ETN66221.1 GGFJ01010233 MBW59374.1 AJWK01025830 CH940648 KQ434869 KZC09237.1 KQ971312 EEZ98628.2 NNAY01001588 OXU23510.1 JR051745 AEY61551.1 KZ288293 PBC29054.1 GG666679 EEN44038.1 GG666487 EEN64219.1 AAZX01005033 LBMM01000423 KMQ98396.1 KK107447 EZA50750.1 KRF79633.1 RCHS01002309 RMX48134.1 JH816866 EKC41731.1 ADTU01027818 CH933808 AAAB01008986 EAA00134.4 APCN01003272 NCKU01000233 RWS16423.1 ATLV01027100 KE525423 KFB53917.1 KK853502 KDR07129.1 CH916367 EDW01111.1 CH963850 DS233727 GAPW01005159 JAC08439.1 KQ982706 KYQ51823.1 KQ976565 KYM80434.1 GL888237 EGI64144.1 GFDL01006285 JAV28760.1 GFDF01002050 JAV12034.1 GDAI01000976 JAI16627.1 AFYH01010809 JXUM01063949 JXUM01063950 KQ562275 KXJ76259.1 KQ977063 KYN05977.1 LSMT01000080 PFX28608.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000007798
+ More
UP000092553 UP000009046 UP000007801 UP000000304 UP000001292 UP000000803 UP000092462 UP000192221 UP000002282 UP000008744 UP000001819 UP000268350 UP000008711 UP000053825 UP000002358 UP000215335 UP000075884 UP000075886 UP000000673 UP000092461 UP000008792 UP000076502 UP000069272 UP000007266 UP000242457 UP000001554 UP000075900 UP000076408 UP000036403 UP000053097 UP000075901 UP000275408 UP000005408 UP000075885 UP000005205 UP000009192 UP000007062 UP000075902 UP000075840 UP000285301 UP000076407 UP000030765 UP000027135 UP000001070 UP000075920 UP000075881 UP000075903 UP000075882 UP000002320 UP000075809 UP000078540 UP000007755 UP000008672 UP000069940 UP000249989 UP000078542 UP000225706
UP000092553 UP000009046 UP000007801 UP000000304 UP000001292 UP000000803 UP000092462 UP000192221 UP000002282 UP000008744 UP000001819 UP000268350 UP000008711 UP000053825 UP000002358 UP000215335 UP000075884 UP000075886 UP000000673 UP000092461 UP000008792 UP000076502 UP000069272 UP000007266 UP000242457 UP000001554 UP000075900 UP000076408 UP000036403 UP000053097 UP000075901 UP000275408 UP000005408 UP000075885 UP000005205 UP000009192 UP000007062 UP000075902 UP000075840 UP000285301 UP000076407 UP000030765 UP000027135 UP000001070 UP000075920 UP000075881 UP000075903 UP000075882 UP000002320 UP000075809 UP000078540 UP000007755 UP000008672 UP000069940 UP000249989 UP000078542 UP000225706
Pfam
PF03937 Sdh5
SUPFAM
SSF109910
SSF109910
Gene 3D
ProteinModelPortal
H9IVF6
I4DPA5
A0A194RGZ9
A0A212FI19
A0A1E1WBP0
A0A2A4JSL7
+ More
A0A2H1VZZ1 B4N665 A0A0M4EHY4 E0VKE1 B3MGU5 A0A0J9TW46 B4QFP7 B4HRL4 A0A1B2AJ40 Q4V5I9 A0A0M3QUL9 A0A1B0DEP3 A0A1W4VPE7 B4P2P8 B4GDB3 B5DZ31 A0A3B0J0G1 B3MI37 A0A2M4ARD1 A0A2M4AMC5 B3N8S9 B4NXN5 A0A0B4KF96 A1Z897 A0A2M4ARF0 B4HMQ1 A0A2M3ZIP8 B5E0U2 A0A0J9R9P0 A0A3B0JM63 H0RNF9 B4QID8 A0A0L7RFV2 K7IQX4 A0A1W4WAK4 B3N6D9 A0A232EUM4 B4GG58 T1EA61 A0A182N346 A0A182QH38 W5JQ07 A0A2M4C275 A0A1B0CSA2 B4LKE5 A0A154PBQ2 A0A182FVN5 D6WAG2 A0A232EYZ0 V9IM23 A0A2A3EC49 C3ZTP0 C3Y5H2 K7J567 A0A182RW68 A0A182YMI9 A0A0J7L6U8 A0A026W3T6 A0A0Q9WCX0 A0A182TCJ5 A0A3M6U3M1 K1S2Y6 A0A182PGE3 A0A158NW55 B4KN44 Q7Q014 A0A182TXH6 A0A182HPU6 A0A3S3P7A0 A0A182WZY2 A0A084WUM3 A0A067QGL4 B4J7I7 A0A182W2A0 A0A182K9U5 B4MRE7 A0A182UN54 A0A182KSX9 B0XK69 A0A023EHW8 A0A151WVC4 A0A195B8A9 F4WNF1 A0A1Q3FMC0 A0A1L8E027 A0A0K8TQI4 H3ABN6 A0A182GHL8 A0A151IM66 A0A2B4SJ61 A0A2K7P6A1
A0A2H1VZZ1 B4N665 A0A0M4EHY4 E0VKE1 B3MGU5 A0A0J9TW46 B4QFP7 B4HRL4 A0A1B2AJ40 Q4V5I9 A0A0M3QUL9 A0A1B0DEP3 A0A1W4VPE7 B4P2P8 B4GDB3 B5DZ31 A0A3B0J0G1 B3MI37 A0A2M4ARD1 A0A2M4AMC5 B3N8S9 B4NXN5 A0A0B4KF96 A1Z897 A0A2M4ARF0 B4HMQ1 A0A2M3ZIP8 B5E0U2 A0A0J9R9P0 A0A3B0JM63 H0RNF9 B4QID8 A0A0L7RFV2 K7IQX4 A0A1W4WAK4 B3N6D9 A0A232EUM4 B4GG58 T1EA61 A0A182N346 A0A182QH38 W5JQ07 A0A2M4C275 A0A1B0CSA2 B4LKE5 A0A154PBQ2 A0A182FVN5 D6WAG2 A0A232EYZ0 V9IM23 A0A2A3EC49 C3ZTP0 C3Y5H2 K7J567 A0A182RW68 A0A182YMI9 A0A0J7L6U8 A0A026W3T6 A0A0Q9WCX0 A0A182TCJ5 A0A3M6U3M1 K1S2Y6 A0A182PGE3 A0A158NW55 B4KN44 Q7Q014 A0A182TXH6 A0A182HPU6 A0A3S3P7A0 A0A182WZY2 A0A084WUM3 A0A067QGL4 B4J7I7 A0A182W2A0 A0A182K9U5 B4MRE7 A0A182UN54 A0A182KSX9 B0XK69 A0A023EHW8 A0A151WVC4 A0A195B8A9 F4WNF1 A0A1Q3FMC0 A0A1L8E027 A0A0K8TQI4 H3ABN6 A0A182GHL8 A0A151IM66 A0A2B4SJ61 A0A2K7P6A1
PDB
2LM4
E-value=9.30365e-13,
Score=171
Ontologies
GO
Topology
Subcellular location
Mitochondrion matrix
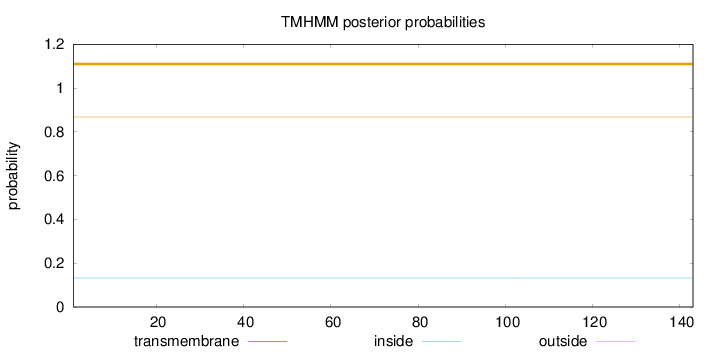
Length:
143
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00102
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.13195
outside
1 - 143
Population Genetic Test Statistics
Pi
295.079968
Theta
203.642063
Tajima's D
1.529089
CLR
0.212268
CSRT
0.793310334483276
Interpretation
Uncertain
