Gene
KWMTBOMO07786
Pre Gene Modal
BGIBMGA000907
Annotation
PREDICTED:_intraflagellar_transport_protein_88_homolog_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.302 Mitochondrial Reliability : 1.442 Nuclear Reliability : 1.172
Sequence
CDS
ATGGTGCGGCTTGGCACGGTAAGCATGCGAGACGTGGGTTCGAGGTCCGGTACATCAGCACGTCCCGTGACCGCACTGCGCGCTGCTGGGTACACCTCGGCGTCACGCGATCGACCCATGCCCACTCAACATTTGGAAGACAGCGTCGAAGATCGCGTTAAGCAAATGGAAGCACGAATCATGGCATTGGTGGAGGAGTCTTGTTTACTGAGCGCCCGACCAGATCCTGATGATGATAGTACTACTAGAAAAGAACATGACTTGGGCCAGGCACTTGCAAAGGCTCAAGAGGCGTCCACGATGGAACGACAGCTTATTCGCATGCAAGAACAAGCGAACTTGGGTGACACTCACAACTTGGATTTGACTTTTGCTGTGCTTTGTAACTTGGCGGATCAGTACGCACTGAACGAGATGTACACTGAAGCACTGAACACGTATCAGTTGCTGACTAGAAACAAGTTGTTTCCGCATGCTAACCGACTCAAGGTGAACATGGGAAATATTTATTTTAAAATGGGAGAACATCCGAAAGCTTTGAAGCTTTATAGGATGGCACTAGATCAAACGCCGACAGCCGAAAAGGATTTGAGAATGAAAGTAATGCATAACATTGGTCTCCTCTTGGTTCGTATGGGAAAATTTCGGGATGCTGTAACAAATTTTCAGCACATAATGCACGAACAAGGTGATTTTCAAACAGGTCTACACCTGGTTTTATGCTCTGTGGCACTCAGTGACGCAGAAGGAGGGAAAGCTGCTTTCCATGCAATGCTGGATGTCGAACCGCCGACTTACCACCAGGACATTCCTATTGATGACGAGAAAGATACTTACGAGTGCGTTGTCCGCGACGTGTCACGCGGAGATAAACTGTCTCGCTGGTCTCGGCGCGCGGCCGCTGAAGCTGAACGATGTCTGGCACTGGCCGCTGCGGTCCTCACACCCACAGCGGGGACGGACGATGATTCAGGGTGA
Protein
MVRLGTVSMRDVGSRSGTSARPVTALRAAGYTSASRDRPMPTQHLEDSVEDRVKQMEARIMALVEESCLLSARPDPDDDSTTRKEHDLGQALAKAQEASTMERQLIRMQEQANLGDTHNLDLTFAVLCNLADQYALNEMYTEALNTYQLLTRNKLFPHANRLKVNMGNIYFKMGEHPKALKLYRMALDQTPTAEKDLRMKVMHNIGLLLVRMGKFRDAVTNFQHIMHEQGDFQTGLHLVLCSVALSDAEGGKAAFHAMLDVEPPTYHQDIPIDDEKDTYECVVRDVSRGDKLSRWSRRAAAEAERCLALAAAVLTPTAGTDDDSG
Summary
Uniprot
A0A2H1V0W0
A0A2A4K4L5
A0A194PDX8
A0A194RHE3
H9IUH7
A0A212FI03
+ More
A0A1B6D4C7 A0A182GCL0 A0A182GTE6 A0A182T0Z5 Q170Z5 A0A0P4VXQ7 A0A2J7QW55 A0A1B6MSL4 A0A182FC49 B0WMM8 W5JMJ3 A0A182K9R2 A0A0K2V4U7 Q5TR18 T1IRH6 A0A182VJN3 A0A182PUS8 A0A084WKC3 A0A182LLC5 A0A182I106 A0A182LWX9 A0A182X0V3 A0A1W4WAB5 A0A158NYM8 A0A182QMI6 A0A0P4VSY1 A0A182WCE8 A0A0P4VZY5 A0A182RD53 A0A182YLD6 A0A182JBM2 V3ZVG1 A0A067R699 A0A182NC56 C3YI17 A0A232ENE1 K7J4M1 A0A1S3HGK4 A0A1S3HGH7 A0A195B1Q2 A0A1B0D291 A0A1W4WAE6 K1QZL1 A0A151WTW7 A0A2T7P3V8 A0A195FTZ5 E2C3N0 E0VSX6 A0A0J7NVH0 E2ANK8 F4WXX6 E9J4T4 A0A336MIV5 A0A1J1IIN9 A0A3R7QAT0 A0A210PHP8 A0A0K8TIU5 A0A026WTW6 A0A154PQV1 R7TWS1 A0A2B4SZ51 D6WHQ7 A7T8G7 A0A0B7BL31 A0A2G8JYD8 A0A2G8K2I0 A0A3L8DVA3 A0A2S2P713 A0A0M8ZXG9 H2ZWI9 A0A2A3ENM9 M3XJE8 D2ILS7 A0A3B4UMY8 V9KG67 V9KG26 A0A088ADN2 I3KF91 A0A3Q3AP65 G3N915 A0A3B4UKZ1 A0A3Q1CSN7 A0A3B4UKD1 A0A3B4YPP8 A0A3P8U430 A0A3B4Y9F9 A0A3P9CUG5 A0A3Q3AQ34 A0A3Q3AQJ6 A0A3P8U457 A0A3Q3B3J0 A0A3B4Z3W6
A0A1B6D4C7 A0A182GCL0 A0A182GTE6 A0A182T0Z5 Q170Z5 A0A0P4VXQ7 A0A2J7QW55 A0A1B6MSL4 A0A182FC49 B0WMM8 W5JMJ3 A0A182K9R2 A0A0K2V4U7 Q5TR18 T1IRH6 A0A182VJN3 A0A182PUS8 A0A084WKC3 A0A182LLC5 A0A182I106 A0A182LWX9 A0A182X0V3 A0A1W4WAB5 A0A158NYM8 A0A182QMI6 A0A0P4VSY1 A0A182WCE8 A0A0P4VZY5 A0A182RD53 A0A182YLD6 A0A182JBM2 V3ZVG1 A0A067R699 A0A182NC56 C3YI17 A0A232ENE1 K7J4M1 A0A1S3HGK4 A0A1S3HGH7 A0A195B1Q2 A0A1B0D291 A0A1W4WAE6 K1QZL1 A0A151WTW7 A0A2T7P3V8 A0A195FTZ5 E2C3N0 E0VSX6 A0A0J7NVH0 E2ANK8 F4WXX6 E9J4T4 A0A336MIV5 A0A1J1IIN9 A0A3R7QAT0 A0A210PHP8 A0A0K8TIU5 A0A026WTW6 A0A154PQV1 R7TWS1 A0A2B4SZ51 D6WHQ7 A7T8G7 A0A0B7BL31 A0A2G8JYD8 A0A2G8K2I0 A0A3L8DVA3 A0A2S2P713 A0A0M8ZXG9 H2ZWI9 A0A2A3ENM9 M3XJE8 D2ILS7 A0A3B4UMY8 V9KG67 V9KG26 A0A088ADN2 I3KF91 A0A3Q3AP65 G3N915 A0A3B4UKZ1 A0A3Q1CSN7 A0A3B4UKD1 A0A3B4YPP8 A0A3P8U430 A0A3B4Y9F9 A0A3P9CUG5 A0A3Q3AQ34 A0A3Q3AQJ6 A0A3P8U457 A0A3Q3B3J0 A0A3B4Z3W6
Pubmed
26354079
19121390
22118469
26483478
17510324
20920257
+ More
23761445 12364791 14747013 17210077 24438588 20966253 21347285 25244985 23254933 24845553 18563158 28648823 20075255 22992520 20798317 20566863 21719571 21282665 28812685 24508170 18362917 19820115 17615350 29023486 30249741 9215903 19933103 24402279 25186727
23761445 12364791 14747013 17210077 24438588 20966253 21347285 25244985 23254933 24845553 18563158 28648823 20075255 22992520 20798317 20566863 21719571 21282665 28812685 24508170 18362917 19820115 17615350 29023486 30249741 9215903 19933103 24402279 25186727
EMBL
ODYU01000170
SOQ34493.1
NWSH01000189
PCG78602.1
KQ459606
KPI91228.1
+ More
KQ460205 KPJ16859.1 BABH01001026 AGBW02008449 OWR53374.1 GEDC01016739 JAS20559.1 JXUM01009771 KQ560291 KXJ83225.1 JXUM01086946 KQ563601 KXJ73605.1 CH477462 EAT40549.1 GDRN01099877 JAI58679.1 NEVH01009768 PNF32806.1 GEBQ01001040 JAT38937.1 DS231999 EDS31110.1 ADMH02000619 ETN65597.1 HACA01027615 CDW44976.1 AAAB01008960 EAL40045.3 JH431361 ATLV01024108 KE525349 KFB50667.1 APCN01000010 AXCM01004546 ADTU01004082 ADTU01004083 AXCN02000408 GDRN01099881 GDRN01099875 JAI58677.1 GDRN01099879 JAI58678.1 KB203357 ESO84911.1 KK852667 KDR18924.1 GG666514 EEN60154.1 NNAY01003168 OXU19875.1 AAZX01007478 KQ976681 KYM78210.1 AJVK01010540 AJVK01010541 AJVK01010542 JH818378 EKC42517.1 KQ982748 KYQ51293.1 PZQS01000006 PVD28091.1 KQ981276 KYN43762.1 GL452328 EFN77453.1 DS235758 EEB16482.1 LBMM01001396 KMQ96390.1 GL441253 EFN64974.1 GL888434 EGI60912.1 GL768119 EFZ12176.1 UFQS01000985 UFQT01000985 SSX08170.1 SSX28287.1 CVRI01000047 CRK98321.1 QCYY01003863 ROT62023.1 NEDP02076687 OWF36001.1 GBRD01000324 JAG65497.1 KK107107 EZA59408.1 KQ435037 KZC14117.1 AMQN01002409 KB309137 ELT95420.1 LSMT01000009 PFX33717.1 KQ971321 EFA00677.2 DS472696 EDO27720.1 HACG01046737 CEK93602.1 MRZV01001087 PIK40752.1 MRZV01000951 PIK42216.1 QOIP01000003 RLU24364.1 GGMR01012586 MBY25205.1 KQ435824 KOX72066.1 AFYH01134842 AFYH01134843 AFYH01134844 AFYH01134845 AFYH01134846 AFYH01134847 AFYH01134848 AFYH01134849 AFYH01134850 KZ288206 PBC33114.1 GQ337482 ACZ64518.1 JW864289 AFO96806.1 JW864235 AFO96752.1 AERX01041975 AERX01041976
KQ460205 KPJ16859.1 BABH01001026 AGBW02008449 OWR53374.1 GEDC01016739 JAS20559.1 JXUM01009771 KQ560291 KXJ83225.1 JXUM01086946 KQ563601 KXJ73605.1 CH477462 EAT40549.1 GDRN01099877 JAI58679.1 NEVH01009768 PNF32806.1 GEBQ01001040 JAT38937.1 DS231999 EDS31110.1 ADMH02000619 ETN65597.1 HACA01027615 CDW44976.1 AAAB01008960 EAL40045.3 JH431361 ATLV01024108 KE525349 KFB50667.1 APCN01000010 AXCM01004546 ADTU01004082 ADTU01004083 AXCN02000408 GDRN01099881 GDRN01099875 JAI58677.1 GDRN01099879 JAI58678.1 KB203357 ESO84911.1 KK852667 KDR18924.1 GG666514 EEN60154.1 NNAY01003168 OXU19875.1 AAZX01007478 KQ976681 KYM78210.1 AJVK01010540 AJVK01010541 AJVK01010542 JH818378 EKC42517.1 KQ982748 KYQ51293.1 PZQS01000006 PVD28091.1 KQ981276 KYN43762.1 GL452328 EFN77453.1 DS235758 EEB16482.1 LBMM01001396 KMQ96390.1 GL441253 EFN64974.1 GL888434 EGI60912.1 GL768119 EFZ12176.1 UFQS01000985 UFQT01000985 SSX08170.1 SSX28287.1 CVRI01000047 CRK98321.1 QCYY01003863 ROT62023.1 NEDP02076687 OWF36001.1 GBRD01000324 JAG65497.1 KK107107 EZA59408.1 KQ435037 KZC14117.1 AMQN01002409 KB309137 ELT95420.1 LSMT01000009 PFX33717.1 KQ971321 EFA00677.2 DS472696 EDO27720.1 HACG01046737 CEK93602.1 MRZV01001087 PIK40752.1 MRZV01000951 PIK42216.1 QOIP01000003 RLU24364.1 GGMR01012586 MBY25205.1 KQ435824 KOX72066.1 AFYH01134842 AFYH01134843 AFYH01134844 AFYH01134845 AFYH01134846 AFYH01134847 AFYH01134848 AFYH01134849 AFYH01134850 KZ288206 PBC33114.1 GQ337482 ACZ64518.1 JW864289 AFO96806.1 JW864235 AFO96752.1 AERX01041975 AERX01041976
Proteomes
UP000218220
UP000053268
UP000053240
UP000005204
UP000007151
UP000069940
+ More
UP000249989 UP000075901 UP000008820 UP000235965 UP000069272 UP000002320 UP000000673 UP000075881 UP000007062 UP000075903 UP000075885 UP000030765 UP000075882 UP000075840 UP000075883 UP000076407 UP000192223 UP000005205 UP000075886 UP000075920 UP000075900 UP000076408 UP000075880 UP000030746 UP000027135 UP000075884 UP000001554 UP000215335 UP000002358 UP000085678 UP000078540 UP000092462 UP000005408 UP000075809 UP000245119 UP000078541 UP000008237 UP000009046 UP000036403 UP000000311 UP000007755 UP000183832 UP000283509 UP000242188 UP000053097 UP000076502 UP000014760 UP000225706 UP000007266 UP000001593 UP000230750 UP000279307 UP000053105 UP000008672 UP000242457 UP000261420 UP000005203 UP000005207 UP000264800 UP000007635 UP000257160 UP000261360 UP000265080 UP000265160 UP000261400
UP000249989 UP000075901 UP000008820 UP000235965 UP000069272 UP000002320 UP000000673 UP000075881 UP000007062 UP000075903 UP000075885 UP000030765 UP000075882 UP000075840 UP000075883 UP000076407 UP000192223 UP000005205 UP000075886 UP000075920 UP000075900 UP000076408 UP000075880 UP000030746 UP000027135 UP000075884 UP000001554 UP000215335 UP000002358 UP000085678 UP000078540 UP000092462 UP000005408 UP000075809 UP000245119 UP000078541 UP000008237 UP000009046 UP000036403 UP000000311 UP000007755 UP000183832 UP000283509 UP000242188 UP000053097 UP000076502 UP000014760 UP000225706 UP000007266 UP000001593 UP000230750 UP000279307 UP000053105 UP000008672 UP000242457 UP000261420 UP000005203 UP000005207 UP000264800 UP000007635 UP000257160 UP000261360 UP000265080 UP000265160 UP000261400
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2H1V0W0
A0A2A4K4L5
A0A194PDX8
A0A194RHE3
H9IUH7
A0A212FI03
+ More
A0A1B6D4C7 A0A182GCL0 A0A182GTE6 A0A182T0Z5 Q170Z5 A0A0P4VXQ7 A0A2J7QW55 A0A1B6MSL4 A0A182FC49 B0WMM8 W5JMJ3 A0A182K9R2 A0A0K2V4U7 Q5TR18 T1IRH6 A0A182VJN3 A0A182PUS8 A0A084WKC3 A0A182LLC5 A0A182I106 A0A182LWX9 A0A182X0V3 A0A1W4WAB5 A0A158NYM8 A0A182QMI6 A0A0P4VSY1 A0A182WCE8 A0A0P4VZY5 A0A182RD53 A0A182YLD6 A0A182JBM2 V3ZVG1 A0A067R699 A0A182NC56 C3YI17 A0A232ENE1 K7J4M1 A0A1S3HGK4 A0A1S3HGH7 A0A195B1Q2 A0A1B0D291 A0A1W4WAE6 K1QZL1 A0A151WTW7 A0A2T7P3V8 A0A195FTZ5 E2C3N0 E0VSX6 A0A0J7NVH0 E2ANK8 F4WXX6 E9J4T4 A0A336MIV5 A0A1J1IIN9 A0A3R7QAT0 A0A210PHP8 A0A0K8TIU5 A0A026WTW6 A0A154PQV1 R7TWS1 A0A2B4SZ51 D6WHQ7 A7T8G7 A0A0B7BL31 A0A2G8JYD8 A0A2G8K2I0 A0A3L8DVA3 A0A2S2P713 A0A0M8ZXG9 H2ZWI9 A0A2A3ENM9 M3XJE8 D2ILS7 A0A3B4UMY8 V9KG67 V9KG26 A0A088ADN2 I3KF91 A0A3Q3AP65 G3N915 A0A3B4UKZ1 A0A3Q1CSN7 A0A3B4UKD1 A0A3B4YPP8 A0A3P8U430 A0A3B4Y9F9 A0A3P9CUG5 A0A3Q3AQ34 A0A3Q3AQJ6 A0A3P8U457 A0A3Q3B3J0 A0A3B4Z3W6
A0A1B6D4C7 A0A182GCL0 A0A182GTE6 A0A182T0Z5 Q170Z5 A0A0P4VXQ7 A0A2J7QW55 A0A1B6MSL4 A0A182FC49 B0WMM8 W5JMJ3 A0A182K9R2 A0A0K2V4U7 Q5TR18 T1IRH6 A0A182VJN3 A0A182PUS8 A0A084WKC3 A0A182LLC5 A0A182I106 A0A182LWX9 A0A182X0V3 A0A1W4WAB5 A0A158NYM8 A0A182QMI6 A0A0P4VSY1 A0A182WCE8 A0A0P4VZY5 A0A182RD53 A0A182YLD6 A0A182JBM2 V3ZVG1 A0A067R699 A0A182NC56 C3YI17 A0A232ENE1 K7J4M1 A0A1S3HGK4 A0A1S3HGH7 A0A195B1Q2 A0A1B0D291 A0A1W4WAE6 K1QZL1 A0A151WTW7 A0A2T7P3V8 A0A195FTZ5 E2C3N0 E0VSX6 A0A0J7NVH0 E2ANK8 F4WXX6 E9J4T4 A0A336MIV5 A0A1J1IIN9 A0A3R7QAT0 A0A210PHP8 A0A0K8TIU5 A0A026WTW6 A0A154PQV1 R7TWS1 A0A2B4SZ51 D6WHQ7 A7T8G7 A0A0B7BL31 A0A2G8JYD8 A0A2G8K2I0 A0A3L8DVA3 A0A2S2P713 A0A0M8ZXG9 H2ZWI9 A0A2A3ENM9 M3XJE8 D2ILS7 A0A3B4UMY8 V9KG67 V9KG26 A0A088ADN2 I3KF91 A0A3Q3AP65 G3N915 A0A3B4UKZ1 A0A3Q1CSN7 A0A3B4UKD1 A0A3B4YPP8 A0A3P8U430 A0A3B4Y9F9 A0A3P9CUG5 A0A3Q3AQ34 A0A3Q3AQJ6 A0A3P8U457 A0A3Q3B3J0 A0A3B4Z3W6
PDB
2WQH
E-value=0.000217433,
Score=105
Ontologies
GO
GO:0031514
GO:1905515
GO:0097730
GO:0005814
GO:0097546
GO:0042073
GO:0036064
GO:0019894
GO:0042462
GO:0007369
GO:0072019
GO:0045494
GO:0005930
GO:0007368
GO:0071482
GO:0045880
GO:0003351
GO:0000132
GO:0060785
GO:0060122
GO:0005515
GO:0004812
GO:0005524
GO:0006418
GO:0004817
GO:0006423
GO:0016021
GO:0022891
GO:0003707
GO:0051537
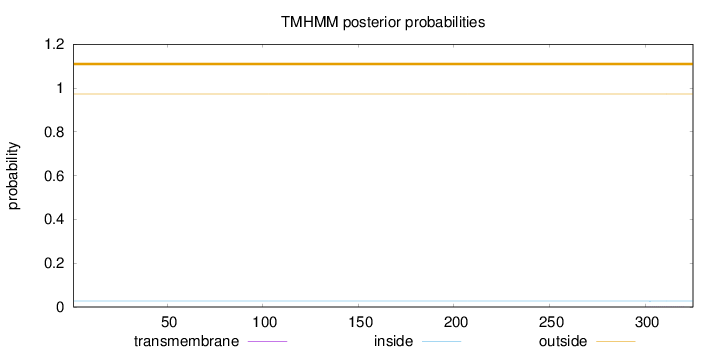
Topology
Length:
325
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00743000000000001
Exp number, first 60 AAs:
0.00043
Total prob of N-in:
0.02728
outside
1 - 325
Population Genetic Test Statistics
Pi
280.369271
Theta
204.243739
Tajima's D
1.353526
CLR
0.249019
CSRT
0.751212439378031
Interpretation
Uncertain
