Gene
KWMTBOMO07785
Pre Gene Modal
BGIBMGA000906
Annotation
PREDICTED:_uncharacterized_protein_LOC101736979_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.121
Sequence
CDS
ATGGTTCTCTTTGTTTGTAAATGTCTTAATGTTTCGTTAGAAAGTGATAAAGTAGAAGATAATGTTGATCGTGAAAAATTGGAGCTTACATCTAGCGAACAAAGGGATATTTTTTTCAGTGAGAAACTCTTATCGTCTCCTGCCAACATATTAAAAACTCAAATAGAACATCAAATTTTAGTTGGTACTAGTGTGGTGGGACGTTGGACTCTACATACTTGCCTGGCCTGTGGAATGACTGTGTATGCTCTATTGCAGGACAATAGAGGATCATCTATTGTATTAATTAACAAACAAATGCAAACAACCCATGAACAAGTAAACTCCCTTAAGAAATCAAATAATTATTCACCAGTCTTTAATATTTTGGTGCCAGAGTTATCTAGTGATATTGAAATGAAGGAGAATATGGATACAAATAATTTTATACTTAATAATAAGAAAAATTGGCCATTAAATAACCAAGTCATTGGTACTCTTAGTAAACAGCTTAATCAAAGGCTCGAATCACAGCTAGAGGCCATCGAAGATACTGTGAGACAGTTTCGAGAACAAAAGTACTCTGAATTTGAAGCTTATAAAGAAAGAGCTCAGAGGGATCATAAAACATTGGCAAGCATAATCAATAAAGCACGAAGTGACAGTGAATGTGAATGGAGAGTTGACACAAGCCTCGATAATGGCCCTCCTTCACCGCAACTACCTCCATTGCAGAGAAGAAGACTATCATCTTTCAAAGATGTCAAAAAAGCTCAGAATATTTTTAAGCCAAACAGAAACCCACCTCAAGAAGATGACTCCTTGGATAATGAAGTTCTTTTTTATTTGGAAGATACGGATTCGCGACACAACATACCATCAGATCAAGATGATTATGAGACTGATGAAGAGAGCGCCGGCGAGGGCGTGCAGATAACTCGGCCCCGCGGCGGAGGCGTTCCCGAGATCGCTCGCTCTCTGCCCATGAACATGCCCAAGTTCCCCAATGACAGAGCGGCACAAGAACAGGATTACGCTGACGATGAGGAGCCTCACGACATAGCGGCCAGCATGAAGGCGTTGGCGCGGTCGGTGCACGGCGACGCGTTCGAGCTGCCGCGGCCGCGCTTCTCCACGCAGATATAG
Protein
MVLFVCKCLNVSLESDKVEDNVDREKLELTSSEQRDIFFSEKLLSSPANILKTQIEHQILVGTSVVGRWTLHTCLACGMTVYALLQDNRGSSIVLINKQMQTTHEQVNSLKKSNNYSPVFNILVPELSSDIEMKENMDTNNFILNNKKNWPLNNQVIGTLSKQLNQRLESQLEAIEDTVRQFREQKYSEFEAYKERAQRDHKTLASIINKARSDSECEWRVDTSLDNGPPSPQLPPLQRRRLSSFKDVKKAQNIFKPNRNPPQEDDSLDNEVLFYLEDTDSRHNIPSDQDDYETDEESAGEGVQITRPRGGGVPEIARSLPMNMPKFPNDRAAQEQDYADDEEPHDIAASMKALARSVHGDAFELPRPRFSTQI
Summary
Uniprot
A0A2A4K3J8
A0A1E1W1Z9
A0A3S2L2M8
A0A2H1V2J8
A0A194RGZ4
A0A194PDA7
+ More
A0A212FI04 A0A0L7LVL5 A0A154PBE9 A0A2A3EDF1 A0A088A9M8 A0A0J7L794 E2A8T4 A0A0L7RFL1 A0A232EH10 K7JYK4 A0A151IIK7 A0A026W422 A0A158NE15 A0A2P8ZFV7 D6X500 A0A1B6GHX9 A0A1Y1K116 A0A151WS17 A0A151JX92 A0A151HZX3 A0A151ISN3 E0VES2 A0A0T6AUY0 F4X445 A0A1B6ECB1 A0A2S2QSH7 E2C492 A0A2S2N9G1 N6TA48 U4U3V7 A0A0K8REV0 B7PFU2 A0A131XX63 A0A1S3IJ09 A0A1L8DDC0 A0A087UKT0 A0A0P4VSZ0 J9K4R1 E9H1F7 A0A023EX62 A0A0L8H2P4 A0A069DRP3 A0A0A9YFJ1 V5GND8 A0A0P5VE14 A0A0P5W861 K1RRN7 A7SHE9 A0A0P4YV30 A0A0P4XY79 A0A293LZK9 A0A0P5Z9X6 A0A0P4YRY6 A0A0P5W0N4 A0A0N8AEN0 A0A0P5N9A5 A0A0P5LNG0 A0A0P4WVP0 A0A0P5Z0R6 A0A0P5Y8B1 A0A0P5UK12 A0A0P5MHM1 A0A0P5U3J0 A0A0N8A2G2 A0A0N8ENW2 C3Z0M8 A0A2B4SD71 A0A023G1V3 A0A023FS54 A0A2R5L7U6 A0A0P5T8M3 A0A131Z3L1 A0A0P4ZZ86 A0A0P5D8M7 L7MFE1 A0A224YF83
A0A212FI04 A0A0L7LVL5 A0A154PBE9 A0A2A3EDF1 A0A088A9M8 A0A0J7L794 E2A8T4 A0A0L7RFL1 A0A232EH10 K7JYK4 A0A151IIK7 A0A026W422 A0A158NE15 A0A2P8ZFV7 D6X500 A0A1B6GHX9 A0A1Y1K116 A0A151WS17 A0A151JX92 A0A151HZX3 A0A151ISN3 E0VES2 A0A0T6AUY0 F4X445 A0A1B6ECB1 A0A2S2QSH7 E2C492 A0A2S2N9G1 N6TA48 U4U3V7 A0A0K8REV0 B7PFU2 A0A131XX63 A0A1S3IJ09 A0A1L8DDC0 A0A087UKT0 A0A0P4VSZ0 J9K4R1 E9H1F7 A0A023EX62 A0A0L8H2P4 A0A069DRP3 A0A0A9YFJ1 V5GND8 A0A0P5VE14 A0A0P5W861 K1RRN7 A7SHE9 A0A0P4YV30 A0A0P4XY79 A0A293LZK9 A0A0P5Z9X6 A0A0P4YRY6 A0A0P5W0N4 A0A0N8AEN0 A0A0P5N9A5 A0A0P5LNG0 A0A0P4WVP0 A0A0P5Z0R6 A0A0P5Y8B1 A0A0P5UK12 A0A0P5MHM1 A0A0P5U3J0 A0A0N8A2G2 A0A0N8ENW2 C3Z0M8 A0A2B4SD71 A0A023G1V3 A0A023FS54 A0A2R5L7U6 A0A0P5T8M3 A0A131Z3L1 A0A0P4ZZ86 A0A0P5D8M7 L7MFE1 A0A224YF83
Pubmed
EMBL
NWSH01000189
PCG78606.1
GDQN01010034
GDQN01005629
JAT81020.1
JAT85425.1
+ More
RSAL01000208 RVE44285.1 ODYU01000170 SOQ34494.1 KQ460205 KPJ16857.1 KQ459606 KPI91227.1 AGBW02008449 OWR53372.1 JTDY01000008 KOB79480.1 KQ434869 KZC09239.1 KZ288293 PBC29051.1 LBMM01000423 KMQ98399.1 GL437663 EFN70163.1 KQ414606 KOC69614.1 NNAY01004629 OXU17635.1 KQ977477 KYN02497.1 KK107447 QOIP01000001 EZA50753.1 RLU26998.1 ADTU01013096 PYGN01000071 PSN55384.1 KQ971381 EEZ97603.1 GECZ01007725 JAS62044.1 GEZM01096065 JAV55174.1 KQ982796 KYQ50603.1 KQ981607 KYN39406.1 KQ976687 KYM78135.1 KQ981064 KYN09839.1 DS235096 EEB11878.1 LJIG01022760 KRT78866.1 GL888625 EGI58769.1 GEDC01001723 JAS35575.1 GGMS01010879 MBY80082.1 GL452434 EFN77225.1 GGMR01001119 MBY13738.1 APGK01038087 KB740952 ENN77129.1 KB632081 ERL88574.1 GADI01004784 JAA69024.1 ABJB010240118 ABJB010260191 ABJB010750753 ABJB011069304 DS704057 EEC05464.1 GEFM01004924 JAP70872.1 GFDF01009707 JAV04377.1 KK120286 KFM77969.1 GDKW01000293 JAI56302.1 ABLF02026352 ABLF02026355 GL732583 EFX74506.1 GBBI01004940 JAC13772.1 KQ419470 KOF83482.1 GBGD01002146 JAC86743.1 GBHO01012665 GDHC01002090 JAG30939.1 JAQ16539.1 GANP01012588 JAB71880.1 GDIP01103150 JAM00565.1 GDIP01103149 JAM00566.1 JH816125 EKC37086.1 DS469659 EDO36904.1 GDIP01233093 JAI90308.1 GDIP01235190 JAI88211.1 GFWV01008757 MAA33486.1 GDIP01059510 LRGB01000024 JAM44205.1 KZS21403.1 GDIP01223386 JAJ00016.1 GDIP01093451 JAM10264.1 GDIP01154897 JAJ68505.1 GDIQ01145390 JAL06336.1 GDIQ01177773 JAK73952.1 GDIP01251435 GDIQ01145389 JAI71966.1 JAL06337.1 GDIP01051021 JAM52694.1 GDIP01061460 JAM42255.1 GDIP01111850 JAL91864.1 GDIQ01158077 JAK93648.1 GDIP01118651 JAL85063.1 GDIP01189041 JAJ34361.1 GDIQ01008372 JAN86365.1 GG666569 EEN53908.1 LSMT01000080 PFX28624.1 GBBL01000332 JAC26988.1 GBBK01000909 JAC23573.1 GGLE01001445 MBY05571.1 GDIQ01095440 JAL56286.1 GEDV01002660 JAP85897.1 GDIP01209390 JAJ14012.1 GDIP01159395 JAJ64007.1 GACK01002108 JAA62926.1 GFPF01001284 MAA12430.1
RSAL01000208 RVE44285.1 ODYU01000170 SOQ34494.1 KQ460205 KPJ16857.1 KQ459606 KPI91227.1 AGBW02008449 OWR53372.1 JTDY01000008 KOB79480.1 KQ434869 KZC09239.1 KZ288293 PBC29051.1 LBMM01000423 KMQ98399.1 GL437663 EFN70163.1 KQ414606 KOC69614.1 NNAY01004629 OXU17635.1 KQ977477 KYN02497.1 KK107447 QOIP01000001 EZA50753.1 RLU26998.1 ADTU01013096 PYGN01000071 PSN55384.1 KQ971381 EEZ97603.1 GECZ01007725 JAS62044.1 GEZM01096065 JAV55174.1 KQ982796 KYQ50603.1 KQ981607 KYN39406.1 KQ976687 KYM78135.1 KQ981064 KYN09839.1 DS235096 EEB11878.1 LJIG01022760 KRT78866.1 GL888625 EGI58769.1 GEDC01001723 JAS35575.1 GGMS01010879 MBY80082.1 GL452434 EFN77225.1 GGMR01001119 MBY13738.1 APGK01038087 KB740952 ENN77129.1 KB632081 ERL88574.1 GADI01004784 JAA69024.1 ABJB010240118 ABJB010260191 ABJB010750753 ABJB011069304 DS704057 EEC05464.1 GEFM01004924 JAP70872.1 GFDF01009707 JAV04377.1 KK120286 KFM77969.1 GDKW01000293 JAI56302.1 ABLF02026352 ABLF02026355 GL732583 EFX74506.1 GBBI01004940 JAC13772.1 KQ419470 KOF83482.1 GBGD01002146 JAC86743.1 GBHO01012665 GDHC01002090 JAG30939.1 JAQ16539.1 GANP01012588 JAB71880.1 GDIP01103150 JAM00565.1 GDIP01103149 JAM00566.1 JH816125 EKC37086.1 DS469659 EDO36904.1 GDIP01233093 JAI90308.1 GDIP01235190 JAI88211.1 GFWV01008757 MAA33486.1 GDIP01059510 LRGB01000024 JAM44205.1 KZS21403.1 GDIP01223386 JAJ00016.1 GDIP01093451 JAM10264.1 GDIP01154897 JAJ68505.1 GDIQ01145390 JAL06336.1 GDIQ01177773 JAK73952.1 GDIP01251435 GDIQ01145389 JAI71966.1 JAL06337.1 GDIP01051021 JAM52694.1 GDIP01061460 JAM42255.1 GDIP01111850 JAL91864.1 GDIQ01158077 JAK93648.1 GDIP01118651 JAL85063.1 GDIP01189041 JAJ34361.1 GDIQ01008372 JAN86365.1 GG666569 EEN53908.1 LSMT01000080 PFX28624.1 GBBL01000332 JAC26988.1 GBBK01000909 JAC23573.1 GGLE01001445 MBY05571.1 GDIQ01095440 JAL56286.1 GEDV01002660 JAP85897.1 GDIP01209390 JAJ14012.1 GDIP01159395 JAJ64007.1 GACK01002108 JAA62926.1 GFPF01001284 MAA12430.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000076502 UP000242457 UP000005203 UP000036403 UP000000311 UP000053825 UP000215335 UP000002358 UP000078542 UP000053097 UP000279307 UP000005205 UP000245037 UP000007266 UP000075809 UP000078541 UP000078540 UP000078492 UP000009046 UP000007755 UP000008237 UP000019118 UP000030742 UP000001555 UP000085678 UP000054359 UP000007819 UP000000305 UP000053454 UP000005408 UP000001593 UP000076858 UP000001554 UP000225706
UP000076502 UP000242457 UP000005203 UP000036403 UP000000311 UP000053825 UP000215335 UP000002358 UP000078542 UP000053097 UP000279307 UP000005205 UP000245037 UP000007266 UP000075809 UP000078541 UP000078540 UP000078492 UP000009046 UP000007755 UP000008237 UP000019118 UP000030742 UP000001555 UP000085678 UP000054359 UP000007819 UP000000305 UP000053454 UP000005408 UP000001593 UP000076858 UP000001554 UP000225706
Pfam
PF15798 PRAS
Interpro
IPR026682
AKT1S1
ProteinModelPortal
A0A2A4K3J8
A0A1E1W1Z9
A0A3S2L2M8
A0A2H1V2J8
A0A194RGZ4
A0A194PDA7
+ More
A0A212FI04 A0A0L7LVL5 A0A154PBE9 A0A2A3EDF1 A0A088A9M8 A0A0J7L794 E2A8T4 A0A0L7RFL1 A0A232EH10 K7JYK4 A0A151IIK7 A0A026W422 A0A158NE15 A0A2P8ZFV7 D6X500 A0A1B6GHX9 A0A1Y1K116 A0A151WS17 A0A151JX92 A0A151HZX3 A0A151ISN3 E0VES2 A0A0T6AUY0 F4X445 A0A1B6ECB1 A0A2S2QSH7 E2C492 A0A2S2N9G1 N6TA48 U4U3V7 A0A0K8REV0 B7PFU2 A0A131XX63 A0A1S3IJ09 A0A1L8DDC0 A0A087UKT0 A0A0P4VSZ0 J9K4R1 E9H1F7 A0A023EX62 A0A0L8H2P4 A0A069DRP3 A0A0A9YFJ1 V5GND8 A0A0P5VE14 A0A0P5W861 K1RRN7 A7SHE9 A0A0P4YV30 A0A0P4XY79 A0A293LZK9 A0A0P5Z9X6 A0A0P4YRY6 A0A0P5W0N4 A0A0N8AEN0 A0A0P5N9A5 A0A0P5LNG0 A0A0P4WVP0 A0A0P5Z0R6 A0A0P5Y8B1 A0A0P5UK12 A0A0P5MHM1 A0A0P5U3J0 A0A0N8A2G2 A0A0N8ENW2 C3Z0M8 A0A2B4SD71 A0A023G1V3 A0A023FS54 A0A2R5L7U6 A0A0P5T8M3 A0A131Z3L1 A0A0P4ZZ86 A0A0P5D8M7 L7MFE1 A0A224YF83
A0A212FI04 A0A0L7LVL5 A0A154PBE9 A0A2A3EDF1 A0A088A9M8 A0A0J7L794 E2A8T4 A0A0L7RFL1 A0A232EH10 K7JYK4 A0A151IIK7 A0A026W422 A0A158NE15 A0A2P8ZFV7 D6X500 A0A1B6GHX9 A0A1Y1K116 A0A151WS17 A0A151JX92 A0A151HZX3 A0A151ISN3 E0VES2 A0A0T6AUY0 F4X445 A0A1B6ECB1 A0A2S2QSH7 E2C492 A0A2S2N9G1 N6TA48 U4U3V7 A0A0K8REV0 B7PFU2 A0A131XX63 A0A1S3IJ09 A0A1L8DDC0 A0A087UKT0 A0A0P4VSZ0 J9K4R1 E9H1F7 A0A023EX62 A0A0L8H2P4 A0A069DRP3 A0A0A9YFJ1 V5GND8 A0A0P5VE14 A0A0P5W861 K1RRN7 A7SHE9 A0A0P4YV30 A0A0P4XY79 A0A293LZK9 A0A0P5Z9X6 A0A0P4YRY6 A0A0P5W0N4 A0A0N8AEN0 A0A0P5N9A5 A0A0P5LNG0 A0A0P4WVP0 A0A0P5Z0R6 A0A0P5Y8B1 A0A0P5UK12 A0A0P5MHM1 A0A0P5U3J0 A0A0N8A2G2 A0A0N8ENW2 C3Z0M8 A0A2B4SD71 A0A023G1V3 A0A023FS54 A0A2R5L7U6 A0A0P5T8M3 A0A131Z3L1 A0A0P4ZZ86 A0A0P5D8M7 L7MFE1 A0A224YF83
Ontologies
PANTHER
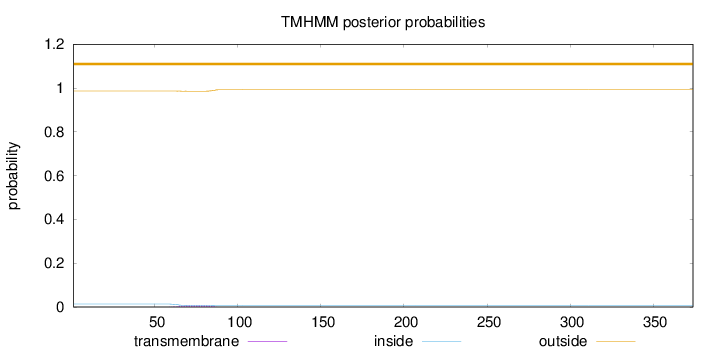
Topology
Length:
374
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15111
Exp number, first 60 AAs:
0.00161
Total prob of N-in:
0.01377
outside
1 - 374
Population Genetic Test Statistics
Pi
198.780814
Theta
155.382555
Tajima's D
0.791632
CLR
0.349065
CSRT
0.604069796510174
Interpretation
Uncertain
