Gene
KWMTBOMO07781
Pre Gene Modal
BGIBMGA000904
Annotation
PREDICTED:_SUN_domain-containing_protein_2-like_[Bombyx_mori]
Full name
SUN domain-containing protein 2
Alternative Name
Protein unc-84 homolog B
Rab5-interacting protein
Sad1/unc-84 protein-like 2
Rab5-interacting protein
Sad1/unc-84 protein-like 2
Location in the cell
Cytoplasmic Reliability : 1.169 Mitochondrial Reliability : 1.087
Sequence
CDS
ATGGAACGGATGTCGGCGGTGATTCCTGCCGTGGCTGCGGCTGCAGGACGCGCTAAAGACGCCTTGGAACCATCTCTGAGGAAGAACAGCCGACAGGCATTGGATAATTATGATTATGATAGACAGGTGGCTGATTACGCGCTCGAGTCTGCTGGTGGTCGCATATTAGATACTGGAGATACAATAGAGCATCTAGTTTATGAGTCGCCCATAAGTTGGGGCTTACATCTAATTACGTCGTGGATGTGTAGGGAATGTCAAGGGGCCAGTGCTATGATCCGCCCAGGTACTTTACCTGGTGAATGCTGGGCGTTCAAAGGATCGAAAGGTCAAGCTATGATACGTCTACTCGGTACTGTGAAGGTGATGGGTGTTAGTGTGGAGCATATTCCGGCGCATATCTCGCCAACCAGGGAAATATCTTCGGCGCCACGACTATTCCAAGTGGAAGGCTTAGAATATCGCAGCGATCCGTACCCCCACGACTTTGGTACCTTTGAGTACGATAAGGAGGGCAAACCGATCCAGTACTTTGAAGTGTTGTACCCTTCAACGAAGGGGTACAGCTTGATAAGGATACGCGTGCTTACTAATTGGGGACATCCTGTGTATACTTGCGTGTACCGAGTGAGGGTTCACGGTGAATTGTCGGGACGTAATCAAAACTTCGGTGCCGACGACACTGAGATGCGAATCGAAAATGAATGA
Protein
MERMSAVIPAVAAAAGRAKDALEPSLRKNSRQALDNYDYDRQVADYALESAGGRILDTGDTIEHLVYESPISWGLHLITSWMCRECQGASAMIRPGTLPGECWAFKGSKGQAMIRLLGTVKVMGVSVEHIPAHISPTREISSAPRLFQVEGLEYRSDPYPHDFGTFEYDKEGKPIQYFEVLYPSTKGYSLIRIRVLTNWGHPVYTCVYRVRVHGELSGRNQNFGADDTEMRIENE
Summary
Description
As a component of the LINC (LInker of Nucleoskeleton and Cytoskeleton) complex, involved in the connection between the nuclear lamina and the cytoskeleton. The nucleocytoplasmic interactions established by the LINC complex play an important role in the transmission of mechanical forces across the nuclear envelope and in nuclear movement and positioning. Specifically, SYNE2 and SUN2 assemble in arrays of transmembrane actin-associated nuclear (TAN) lines which are bound to F-actin cables and couple the nucleus to retrograde actin flow during actin-dependent nuclear movement. Required for interkinetic nuclear migration (INM) and essential for nucleokinesis and centrosome-nucleus coupling during radial neuronal migration in the cerebral cortex and during glial migration. Required for nuclear migration in retinal photoreceptor progenitors implicating association with cytoplasmic dynein-dynactin and kinesin motor complexes, and probably B-type lamins; SUN1 and SUN2 seem to act redundantly. The SUN1/2:KASH5 LINC complex couples telomeres to microtubules during meiosis; SUN1 and SUN2 seem to act at least partial redundantly. Anchors chromosome movement in the prophase of meiosis and is involved in selective gene expression of coding and non-coding RNAs needed for gametogenesis. Required for telomere attachment to nuclear envelope and gametogenesis. May also function on endocytic vesicles as a receptor for RAB5-GDP and participate in the activation of RAB5.
Subunit
Core component of the LINC complex which is composed of inner nuclear membrane SUN domain-containing proteins coupled to outer nuclear membrane KASH domain-containing nesprins. SUN and KASH domain-containing proteins seem to bind each other promiscuously; however, differentially expression of LINC complex constituents is giving rise to specific assemblies. At least SUN1/2-containing core LINC complexes are proposed to be hexameric composed of three protomers of each KASH and SUN domain-containing protein. Interacts with SYNE2; the SUN2:SYNE2/KASH2 LINC complex is a heterohexamer; the homotrimeric cloverleave-like conformation of the SUN domain is a prerequisite for LINC complex formation in which three separate SYNE2/KASH2 peptides bind at the interface of adjacent SUN domains. Component of a probable SUN2:KASH5 LINC complex. Interacts with SYNE1 and SYNE3; probably forming respective LINC complexes. Interacts with A-type lamin. Interaction with lamins B1 and C is hardly detectable (By similarity). Interacts with EMD and RAB5A. Interacts with TMEM43 and TMEM201.
Keywords
3D-structure
Alternative splicing
Coiled coil
Complete proteome
Disulfide bond
Endosome
Glycoprotein
Meiosis
Membrane
Nucleus
Phosphoprotein
Polymorphism
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain SUN domain-containing protein 2
splice variant In isoform 2.
sequence variant In dbSNP:rs2072799.
splice variant In isoform 2.
sequence variant In dbSNP:rs2072799.
Uniprot
H9IUH4
A0A2H1VD83
A0A2A4K3H9
A0A212FI43
A0A194RHD8
A0A194PEV0
+ More
A0A0L7LVD4 A0A151WZS1 A0A195D511 A0A195F5Z7 A0A2A3E2Q6 A0A195BU98 A0A026W149 A0A195EK48 A0A3L8DPI7 A0A0L7RA72 A0A1Y1LL45 A0A154P7G3 A0A088ATM6 F4WXT4 A0A3R7PZ44 K7IWR4 A0A0N5DHZ0 A0A0K2TYC1 A0A1D2MFR8 W5LYM1 Q6NT72 A0A2K5JCU8 H9FSI8 I0FQL1 A0A2Y9SW70 A0A2K6AWC8 A0A2K6QK73 A0A2K6JUX1 A0A2K6JUY5 F7FC46 A0A2K5YQ49 A0A2K5UID6 A0A0D9R2D4 H9Z7D3 A0A2K5UIF2 A0A2K5YQ22 I2CYJ6 L5KFV6 A0A2R9BY76 A0A2I3TT81 A0A2I2ZZ03 A0A024R1P7 Q9UH99 G1RYX5 A0A2I2YDX4 A0A2J8UWP8 A0A2J8LNL1 A0A2I3G549 H2R4A1 A0A096NK39 A0A2R9BP74 Q9UH99-2 A0A2J8UWN1 A0A2K6TID2 A0A2K6TIF2 B4E2A6 A0A232EG81 U3DHD4 U3FQT8 A0A2J7RGG7 A0A2K5R8B3 H0V6Q8 A0A091E028 B4DIU6 A0A2K5R883 A0A2K6TIA0 A0A1D2N6Q4 A0A093BDN0 A0A1S3FWD8 A0A0S7ET58 A0A1S3FUU3 A0A2K5NL97 A0A2U4C0T8 A0A2K5CW18 A0A2Y9QB58 F7B1Y2 A0A2Y9QBA5 A0A346QRB9 A8K129 A0A1U7UNM7 A0A067QSG5 A0A2F0B730 W5KNM3 A6QLV1 A0A250YKY5 K9ITX2 A0A2K6F7S0 A0A0A9YR80 H0WGK7 K9IU18 A0A2I0USJ5 A0A2Y9T081 A0A286ZN31
A0A0L7LVD4 A0A151WZS1 A0A195D511 A0A195F5Z7 A0A2A3E2Q6 A0A195BU98 A0A026W149 A0A195EK48 A0A3L8DPI7 A0A0L7RA72 A0A1Y1LL45 A0A154P7G3 A0A088ATM6 F4WXT4 A0A3R7PZ44 K7IWR4 A0A0N5DHZ0 A0A0K2TYC1 A0A1D2MFR8 W5LYM1 Q6NT72 A0A2K5JCU8 H9FSI8 I0FQL1 A0A2Y9SW70 A0A2K6AWC8 A0A2K6QK73 A0A2K6JUX1 A0A2K6JUY5 F7FC46 A0A2K5YQ49 A0A2K5UID6 A0A0D9R2D4 H9Z7D3 A0A2K5UIF2 A0A2K5YQ22 I2CYJ6 L5KFV6 A0A2R9BY76 A0A2I3TT81 A0A2I2ZZ03 A0A024R1P7 Q9UH99 G1RYX5 A0A2I2YDX4 A0A2J8UWP8 A0A2J8LNL1 A0A2I3G549 H2R4A1 A0A096NK39 A0A2R9BP74 Q9UH99-2 A0A2J8UWN1 A0A2K6TID2 A0A2K6TIF2 B4E2A6 A0A232EG81 U3DHD4 U3FQT8 A0A2J7RGG7 A0A2K5R8B3 H0V6Q8 A0A091E028 B4DIU6 A0A2K5R883 A0A2K6TIA0 A0A1D2N6Q4 A0A093BDN0 A0A1S3FWD8 A0A0S7ET58 A0A1S3FUU3 A0A2K5NL97 A0A2U4C0T8 A0A2K5CW18 A0A2Y9QB58 F7B1Y2 A0A2Y9QBA5 A0A346QRB9 A8K129 A0A1U7UNM7 A0A067QSG5 A0A2F0B730 W5KNM3 A6QLV1 A0A250YKY5 K9ITX2 A0A2K6F7S0 A0A0A9YR80 H0WGK7 K9IU18 A0A2I0USJ5 A0A2Y9T081 A0A286ZN31
Pubmed
19121390
22118469
26354079
26227816
24508170
30249741
+ More
28004739 21719571 20075255 27289101 15489334 25319552 25362486 17431167 23258410 22722832 15164055 16136131 22398555 11181995 10818110 17132086 9734811 15461802 17974005 10591208 10375507 12393179 12239280 12958361 15082709 17081983 17724119 18845190 18396275 19159218 19690332 21391237 21269460 23186163 24275569 26244732 22632968 22945352 22170055 28648823 25243066 21993624 24845553 25329095 19393038 28087693 25401762
28004739 21719571 20075255 27289101 15489334 25319552 25362486 17431167 23258410 22722832 15164055 16136131 22398555 11181995 10818110 17132086 9734811 15461802 17974005 10591208 10375507 12393179 12239280 12958361 15082709 17081983 17724119 18845190 18396275 19159218 19690332 21391237 21269460 23186163 24275569 26244732 22632968 22945352 22170055 28648823 25243066 21993624 24845553 25329095 19393038 28087693 25401762
EMBL
BABH01001039
ODYU01001917
SOQ38788.1
NWSH01000189
PCG78609.1
AGBW02008449
+ More
OWR53369.1 KQ460205 KPJ16854.1 KQ459606 KPI91224.1 JTDY01000008 KOB79483.1 KQ982649 KYQ53201.1 KQ976842 KYN07936.1 KQ981805 KYN35494.1 KZ288452 PBC25546.1 KQ976403 KYM92189.1 KK107570 EZA48789.1 KQ978801 KYN28239.1 QOIP01000005 RLU22317.1 KQ414619 KOC67782.1 GEZM01056552 JAV72655.1 KQ434823 KZC07274.1 GL888427 EGI61012.1 QCYY01003330 ROT64082.1 AAZX01000616 HACA01013663 CDW31024.1 LJIJ01001441 ODM91732.1 AHAT01030100 BC069253 AAH69253.1 JU333841 JU333842 AFE77597.1 JV046666 JV046667 AFI36737.1 JSUE03005524 AQIA01006974 AQIB01136553 JU475045 JU475046 AFH31849.1 JV667368 AFJ72031.1 KB030755 ELK10222.1 AJFE02074311 AJFE02074312 AACZ04068045 CH471095 EAW60270.1 AB014568 AY682988 CR456474 BX537962 AL008583 AL021806 AL021707 BC030684 BC094797 BC111549 BC111717 AF202723 ADFV01024598 ADFV01024599 ABGA01111555 ABGA01111556 ABGA01111557 NDHI03003442 PNJ49677.1 PNJ49678.1 PNJ49680.1 NBAG03000283 PNI48857.1 GABC01000046 GABC01000045 GABF01001739 GABF01001738 GABD01000027 GABD01000026 GABE01000160 GABE01000159 GABE01000158 JAA11292.1 JAA11293.1 JAA20406.1 JAA33073.1 JAA44579.1 PNI48855.1 PNI48858.1 PNI48859.1 AHZZ02001248 PNJ49679.1 AK304188 BAG65068.1 NNAY01004848 OXU17344.1 GAMS01010345 GAMQ01006343 JAB12791.1 JAB35508.1 GAMT01000771 GAMR01009247 GAMP01003994 JAB11090.1 JAB24685.1 JAB48761.1 NEVH01003784 PNF39922.1 AAKN02031258 KN121328 KFO36672.1 AK295787 BAG58608.1 LJIJ01000180 ODN00948.1 KN125643 KFU83601.1 GBYX01474144 GBYX01474141 JAO07516.1 MH156453 AXR98509.1 AK289744 BAF82433.1 KK853076 KDR11726.1 NTJE010088800 MBV98801.1 BC148095 AAI48096.1 GFFW01000602 JAV44186.1 GABZ01000822 JAA52703.1 GBHO01008042 JAG35562.1 AAQR03016211 GABZ01000762 JAA52763.1 KZ505643 PKU49025.1 AEMK02000027
OWR53369.1 KQ460205 KPJ16854.1 KQ459606 KPI91224.1 JTDY01000008 KOB79483.1 KQ982649 KYQ53201.1 KQ976842 KYN07936.1 KQ981805 KYN35494.1 KZ288452 PBC25546.1 KQ976403 KYM92189.1 KK107570 EZA48789.1 KQ978801 KYN28239.1 QOIP01000005 RLU22317.1 KQ414619 KOC67782.1 GEZM01056552 JAV72655.1 KQ434823 KZC07274.1 GL888427 EGI61012.1 QCYY01003330 ROT64082.1 AAZX01000616 HACA01013663 CDW31024.1 LJIJ01001441 ODM91732.1 AHAT01030100 BC069253 AAH69253.1 JU333841 JU333842 AFE77597.1 JV046666 JV046667 AFI36737.1 JSUE03005524 AQIA01006974 AQIB01136553 JU475045 JU475046 AFH31849.1 JV667368 AFJ72031.1 KB030755 ELK10222.1 AJFE02074311 AJFE02074312 AACZ04068045 CH471095 EAW60270.1 AB014568 AY682988 CR456474 BX537962 AL008583 AL021806 AL021707 BC030684 BC094797 BC111549 BC111717 AF202723 ADFV01024598 ADFV01024599 ABGA01111555 ABGA01111556 ABGA01111557 NDHI03003442 PNJ49677.1 PNJ49678.1 PNJ49680.1 NBAG03000283 PNI48857.1 GABC01000046 GABC01000045 GABF01001739 GABF01001738 GABD01000027 GABD01000026 GABE01000160 GABE01000159 GABE01000158 JAA11292.1 JAA11293.1 JAA20406.1 JAA33073.1 JAA44579.1 PNI48855.1 PNI48858.1 PNI48859.1 AHZZ02001248 PNJ49679.1 AK304188 BAG65068.1 NNAY01004848 OXU17344.1 GAMS01010345 GAMQ01006343 JAB12791.1 JAB35508.1 GAMT01000771 GAMR01009247 GAMP01003994 JAB11090.1 JAB24685.1 JAB48761.1 NEVH01003784 PNF39922.1 AAKN02031258 KN121328 KFO36672.1 AK295787 BAG58608.1 LJIJ01000180 ODN00948.1 KN125643 KFU83601.1 GBYX01474144 GBYX01474141 JAO07516.1 MH156453 AXR98509.1 AK289744 BAF82433.1 KK853076 KDR11726.1 NTJE010088800 MBV98801.1 BC148095 AAI48096.1 GFFW01000602 JAV44186.1 GABZ01000822 JAA52703.1 GBHO01008042 JAG35562.1 AAQR03016211 GABZ01000762 JAA52763.1 KZ505643 PKU49025.1 AEMK02000027
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000075809 UP000078542 UP000078541 UP000242457 UP000078540 UP000053097 UP000078492 UP000279307 UP000053825 UP000076502 UP000005203 UP000007755 UP000283509 UP000002358 UP000046395 UP000094527 UP000018468 UP000233080 UP000233120 UP000233200 UP000233180 UP000006718 UP000233140 UP000233100 UP000029965 UP000010552 UP000240080 UP000002277 UP000001519 UP000005640 UP000001073 UP000001595 UP000028761 UP000233220 UP000215335 UP000235965 UP000233040 UP000005447 UP000028990 UP000081671 UP000233060 UP000245320 UP000233020 UP000248483 UP000008225 UP000189704 UP000027135 UP000018467 UP000009136 UP000233160 UP000005225 UP000008227
UP000075809 UP000078542 UP000078541 UP000242457 UP000078540 UP000053097 UP000078492 UP000279307 UP000053825 UP000076502 UP000005203 UP000007755 UP000283509 UP000002358 UP000046395 UP000094527 UP000018468 UP000233080 UP000233120 UP000233200 UP000233180 UP000006718 UP000233140 UP000233100 UP000029965 UP000010552 UP000240080 UP000002277 UP000001519 UP000005640 UP000001073 UP000001595 UP000028761 UP000233220 UP000215335 UP000235965 UP000233040 UP000005447 UP000028990 UP000081671 UP000233060 UP000245320 UP000233020 UP000248483 UP000008225 UP000189704 UP000027135 UP000018467 UP000009136 UP000233160 UP000005225 UP000008227
Interpro
Gene 3D
ProteinModelPortal
H9IUH4
A0A2H1VD83
A0A2A4K3H9
A0A212FI43
A0A194RHD8
A0A194PEV0
+ More
A0A0L7LVD4 A0A151WZS1 A0A195D511 A0A195F5Z7 A0A2A3E2Q6 A0A195BU98 A0A026W149 A0A195EK48 A0A3L8DPI7 A0A0L7RA72 A0A1Y1LL45 A0A154P7G3 A0A088ATM6 F4WXT4 A0A3R7PZ44 K7IWR4 A0A0N5DHZ0 A0A0K2TYC1 A0A1D2MFR8 W5LYM1 Q6NT72 A0A2K5JCU8 H9FSI8 I0FQL1 A0A2Y9SW70 A0A2K6AWC8 A0A2K6QK73 A0A2K6JUX1 A0A2K6JUY5 F7FC46 A0A2K5YQ49 A0A2K5UID6 A0A0D9R2D4 H9Z7D3 A0A2K5UIF2 A0A2K5YQ22 I2CYJ6 L5KFV6 A0A2R9BY76 A0A2I3TT81 A0A2I2ZZ03 A0A024R1P7 Q9UH99 G1RYX5 A0A2I2YDX4 A0A2J8UWP8 A0A2J8LNL1 A0A2I3G549 H2R4A1 A0A096NK39 A0A2R9BP74 Q9UH99-2 A0A2J8UWN1 A0A2K6TID2 A0A2K6TIF2 B4E2A6 A0A232EG81 U3DHD4 U3FQT8 A0A2J7RGG7 A0A2K5R8B3 H0V6Q8 A0A091E028 B4DIU6 A0A2K5R883 A0A2K6TIA0 A0A1D2N6Q4 A0A093BDN0 A0A1S3FWD8 A0A0S7ET58 A0A1S3FUU3 A0A2K5NL97 A0A2U4C0T8 A0A2K5CW18 A0A2Y9QB58 F7B1Y2 A0A2Y9QBA5 A0A346QRB9 A8K129 A0A1U7UNM7 A0A067QSG5 A0A2F0B730 W5KNM3 A6QLV1 A0A250YKY5 K9ITX2 A0A2K6F7S0 A0A0A9YR80 H0WGK7 K9IU18 A0A2I0USJ5 A0A2Y9T081 A0A286ZN31
A0A0L7LVD4 A0A151WZS1 A0A195D511 A0A195F5Z7 A0A2A3E2Q6 A0A195BU98 A0A026W149 A0A195EK48 A0A3L8DPI7 A0A0L7RA72 A0A1Y1LL45 A0A154P7G3 A0A088ATM6 F4WXT4 A0A3R7PZ44 K7IWR4 A0A0N5DHZ0 A0A0K2TYC1 A0A1D2MFR8 W5LYM1 Q6NT72 A0A2K5JCU8 H9FSI8 I0FQL1 A0A2Y9SW70 A0A2K6AWC8 A0A2K6QK73 A0A2K6JUX1 A0A2K6JUY5 F7FC46 A0A2K5YQ49 A0A2K5UID6 A0A0D9R2D4 H9Z7D3 A0A2K5UIF2 A0A2K5YQ22 I2CYJ6 L5KFV6 A0A2R9BY76 A0A2I3TT81 A0A2I2ZZ03 A0A024R1P7 Q9UH99 G1RYX5 A0A2I2YDX4 A0A2J8UWP8 A0A2J8LNL1 A0A2I3G549 H2R4A1 A0A096NK39 A0A2R9BP74 Q9UH99-2 A0A2J8UWN1 A0A2K6TID2 A0A2K6TIF2 B4E2A6 A0A232EG81 U3DHD4 U3FQT8 A0A2J7RGG7 A0A2K5R8B3 H0V6Q8 A0A091E028 B4DIU6 A0A2K5R883 A0A2K6TIA0 A0A1D2N6Q4 A0A093BDN0 A0A1S3FWD8 A0A0S7ET58 A0A1S3FUU3 A0A2K5NL97 A0A2U4C0T8 A0A2K5CW18 A0A2Y9QB58 F7B1Y2 A0A2Y9QBA5 A0A346QRB9 A8K129 A0A1U7UNM7 A0A067QSG5 A0A2F0B730 W5KNM3 A6QLV1 A0A250YKY5 K9ITX2 A0A2K6F7S0 A0A0A9YR80 H0WGK7 K9IU18 A0A2I0USJ5 A0A2Y9T081 A0A286ZN31
PDB
3UNP
E-value=1.59727e-34,
Score=363
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Nucleus inner membrane
Nucleus envelope
Endosome membrane
Nucleus envelope
Endosome membrane
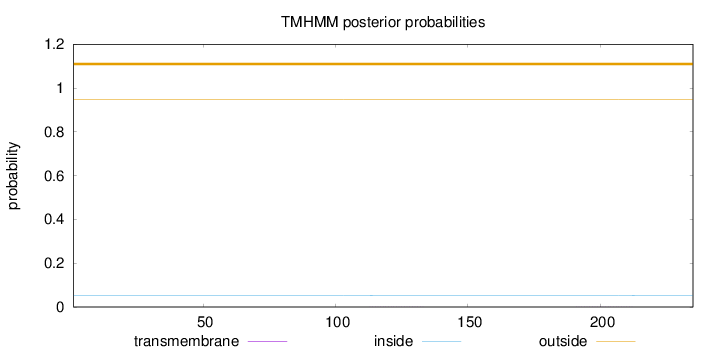
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01067
Exp number, first 60 AAs:
0.00031
Total prob of N-in:
0.05231
outside
1 - 235
Population Genetic Test Statistics
Pi
240.076015
Theta
152.77694
Tajima's D
1.351752
CLR
1.694449
CSRT
0.753512324383781
Interpretation
Uncertain
