Gene
KWMTBOMO07778
Pre Gene Modal
BGIBMGA001240
Annotation
PREDICTED:_CD151_antigen_[Bombyx_mori]
Full name
Tetraspanin
+ More
Multifunctional fusion protein
CTP synthase
Multifunctional fusion protein
CTP synthase
Alternative Name
UTP--ammonia ligase
Location in the cell
PlasmaMembrane Reliability : 3.453
Sequence
CDS
ATGGCAGCAGGCCTCACCATGTGTGCGACGGCTGCTCTTGGCTGCTGGGCAACCTATTTACCCGGATATGCCGCTTTGAGTATCTACTTCTTAATAGTTCTGTCTTTGCTATTGTGCGAGTGTGCTGGAGGCGTGTTAGCTGCAGTATGGCCGCGATGTCTGGGACTTGAAAACGCACGAGGGGGCTCTGTCGGAGCTCTGCAAAGCTATTATGCTATGCCCGACTTTGAACAATTCACTGCGTCTGTAGATTTAGCGCAAACAGAGTTGCAGTGTTGCGGTATGACAGATGCTCGAAACTACGACATGTCCGTATGGCAACTGAGGAGGCTGGGTCCGCGCGGAATGGCCGTACCGTTGAGTTGCTGCGTGCAAATCGAAGAGAACATCTCGTATCTGAACCCGATGCCGGTCAATTTATCGCGCTGTCAGGAGTATCAACCTAACCCTGTTTATCGACATGTTCCGGGCTGTATCGGTAAACTTGAAGAGTGGTACCAGAAACAATATTTTGTCCTCATGCTGAGCATCTTCATATTTGCTGTATTCAAACTGGGAGTTTTGTTAAGCACTGTATTTTCATGTATCCGACTCCGTCAAAGGAGACAAGTACTACATACAGTTACAGTTAAATCCATTGATCACGCAACGAATGAAAATATGTACGGTTCGGGAGTTCGCGAAGACCACATAACAGCCAAATATATCCAGCCGGATAATTATTACAGTCCAAGAGTTCGAAACCCTCGTATTTTTACGAGCAAACCGAACGAAATTATATGA
Protein
MAAGLTMCATAALGCWATYLPGYAALSIYFLIVLSLLLCECAGGVLAAVWPRCLGLENARGGSVGALQSYYAMPDFEQFTASVDLAQTELQCCGMTDARNYDMSVWQLRRLGPRGMAVPLSCCVQIEENISYLNPMPVNLSRCQEYQPNPVYRHVPGCIGKLEEWYQKQYFVLMLSIFIFAVFKLGVLLSTVFSCIRLRQRRQVLHTVTVKSIDHATNENMYGSGVREDHITAKYIQPDNYYSPRVRNPRIFTSKPNEII
Summary
Description
Catalyzes the ATP-dependent amination of UTP to CTP with either L-glutamine or ammonia as the source of nitrogen.
Catalytic Activity
ATP + H2O + L-glutamine + UTP = ADP + CTP + 2 H(+) + L-glutamate + phosphate
Similarity
Belongs to the tetraspanin (TM4SF) family.
Belongs to the CTP synthase family.
Belongs to the CTP synthase family.
Uniprot
H9IVG0
A0A2A4K4M5
A0A2H1VDC6
A0A194PD23
A0A3S2NZD4
A0A194RLJ0
+ More
A0A212FI16 A0A0L7LVL0 A0A182SEV8 A0A182W3P9 A0A023EQE6 A0A182YLZ4 A0A182RRD9 Q17D34 A0A182LN67 A0A182XAZ4 Q7QDT4 A0A182HX67 A0A1Q3FGF4 A0A182QPI8 A0A182NDI4 A0A182FM21 U5EX76 A0A084VTR7 A0A182IJ08 A0A182JWN9 A0A336MHF1 B0WVJ3 A0A182PUI8 D6W8H3 A0A1J1IIN1 A0A336KY22 A0A0M4E8N6 A0A0J9UKN3 A0A1W4XDW9 A0A1I8Q7V8 A0A1Y1KBZ6 A0A0L0C3W9 A0A2A3EKG1 U4UB82 A0A0A1WZW0 B4HH84 A0A088A4X2 B4PHW3 A0A0T6B8L0 A0A1A9XX71 B3MAN0 A0A1I8M1X7 A0A1B0AUW5 B4LNS3 A0A1W4UI57 A0A1A9Z0H7 B3NHH4 Q8IQK9 A0A1A9V2R0 Q4V3R4 A0A026W585 A0A0R3NNW6 A0A1B0FCD9 B4GI53 B5DZ48 A0A3L8DQX7 A0A0R3NTN9 B4KT74 B4MYH1 A0A3B0JAP4 B4JVI3 E2A5E6 A0A0C9R1H1 A0A0L7R5M4 E2BB46 A0A232FFR5 K7J7Z6 A0A0M8ZU68 A0A151JT85 A0A158NY22 A0A182UE64 A0A195ECI0 A0A151I588 N6UE05 A0A1S4EMK9 E0VHY1 A0A3Q0JH04 N6TR85 A0A226E8K7 A0A2P6L519
A0A212FI16 A0A0L7LVL0 A0A182SEV8 A0A182W3P9 A0A023EQE6 A0A182YLZ4 A0A182RRD9 Q17D34 A0A182LN67 A0A182XAZ4 Q7QDT4 A0A182HX67 A0A1Q3FGF4 A0A182QPI8 A0A182NDI4 A0A182FM21 U5EX76 A0A084VTR7 A0A182IJ08 A0A182JWN9 A0A336MHF1 B0WVJ3 A0A182PUI8 D6W8H3 A0A1J1IIN1 A0A336KY22 A0A0M4E8N6 A0A0J9UKN3 A0A1W4XDW9 A0A1I8Q7V8 A0A1Y1KBZ6 A0A0L0C3W9 A0A2A3EKG1 U4UB82 A0A0A1WZW0 B4HH84 A0A088A4X2 B4PHW3 A0A0T6B8L0 A0A1A9XX71 B3MAN0 A0A1I8M1X7 A0A1B0AUW5 B4LNS3 A0A1W4UI57 A0A1A9Z0H7 B3NHH4 Q8IQK9 A0A1A9V2R0 Q4V3R4 A0A026W585 A0A0R3NNW6 A0A1B0FCD9 B4GI53 B5DZ48 A0A3L8DQX7 A0A0R3NTN9 B4KT74 B4MYH1 A0A3B0JAP4 B4JVI3 E2A5E6 A0A0C9R1H1 A0A0L7R5M4 E2BB46 A0A232FFR5 K7J7Z6 A0A0M8ZU68 A0A151JT85 A0A158NY22 A0A182UE64 A0A195ECI0 A0A151I588 N6UE05 A0A1S4EMK9 E0VHY1 A0A3Q0JH04 N6TR85 A0A226E8K7 A0A2P6L519
EC Number
6.3.4.2
Pubmed
19121390
26354079
22118469
26227816
24945155
26483478
+ More
25244985 17510324 20966253 12364791 14747013 17210077 24438588 18362917 19820115 22936249 28004739 26108605 23537049 25830018 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 15632085 30249741 20798317 28648823 20075255 21347285 20566863
25244985 17510324 20966253 12364791 14747013 17210077 24438588 18362917 19820115 22936249 28004739 26108605 23537049 25830018 17994087 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 15632085 30249741 20798317 28648823 20075255 21347285 20566863
EMBL
BABH01001040
NWSH01000189
PCG78612.1
ODYU01001917
SOQ38791.1
KQ459606
+ More
KPI91221.1 RSAL01000084 RVE48418.1 KQ460205 KPJ16851.1 AGBW02008449 OWR53366.1 JTDY01000008 KOB79475.1 JXUM01027703 JXUM01027704 JXUM01027705 GAPW01002280 GAPW01002279 KQ560798 JAC11318.1 KXJ80857.1 CH477300 EAT44287.1 AAAB01008849 EAA07108.4 APCN01005145 GFDL01008416 JAV26629.1 AXCN02001759 GANO01000200 JAB59671.1 ATLV01016453 KE525091 KFB41361.1 UFQT01001277 SSX29824.1 DS232127 EDS35600.1 KQ971312 EEZ98301.1 CVRI01000054 CRL00048.1 UFQS01001006 UFQT01001006 SSX08503.1 SSX28419.1 CP012524 ALC41121.1 CM002912 KMY99465.1 GEZM01086831 JAV58969.1 JRES01001007 KNC26189.1 KZ288219 PBC32217.1 KB632277 ERL91179.1 GBXI01009925 JAD04367.1 CH480815 EDW41409.1 CM000159 EDW94438.1 LJIG01009146 KRT83664.1 CH902618 EDV41317.1 JXJN01003777 JXJN01003778 CH940648 EDW60143.1 CH954178 EDV51700.1 AE014296 AAN11811.1 AAN11812.2 BT023292 AAY55708.1 KK107413 EZA51158.1 CM000071 KRT02492.1 CCAG010007972 CH479183 EDW36173.1 EDY69147.1 QOIP01000005 RLU22850.1 KRT02493.1 CH933808 EDW10586.1 CH963894 EDW77160.1 OUUW01000004 SPP79394.1 CH916375 EDV98451.1 GL436923 EFN71338.1 GBYB01001800 JAG71567.1 KQ414648 KOC66187.1 GL446901 EFN87137.1 NNAY01000312 OXU29298.1 KQ435844 KOX71240.1 KQ981972 KYN31634.1 ADTU01002930 KQ979074 KYN22816.1 KQ976434 KYM87159.1 APGK01026367 APGK01026368 KB740627 ENN79940.1 DS235172 EEB12987.1 APGK01008292 APGK01008293 KB737656 ENN82984.1 LNIX01000006 OXA53201.1 MWRG01001773 PRD33677.1
KPI91221.1 RSAL01000084 RVE48418.1 KQ460205 KPJ16851.1 AGBW02008449 OWR53366.1 JTDY01000008 KOB79475.1 JXUM01027703 JXUM01027704 JXUM01027705 GAPW01002280 GAPW01002279 KQ560798 JAC11318.1 KXJ80857.1 CH477300 EAT44287.1 AAAB01008849 EAA07108.4 APCN01005145 GFDL01008416 JAV26629.1 AXCN02001759 GANO01000200 JAB59671.1 ATLV01016453 KE525091 KFB41361.1 UFQT01001277 SSX29824.1 DS232127 EDS35600.1 KQ971312 EEZ98301.1 CVRI01000054 CRL00048.1 UFQS01001006 UFQT01001006 SSX08503.1 SSX28419.1 CP012524 ALC41121.1 CM002912 KMY99465.1 GEZM01086831 JAV58969.1 JRES01001007 KNC26189.1 KZ288219 PBC32217.1 KB632277 ERL91179.1 GBXI01009925 JAD04367.1 CH480815 EDW41409.1 CM000159 EDW94438.1 LJIG01009146 KRT83664.1 CH902618 EDV41317.1 JXJN01003777 JXJN01003778 CH940648 EDW60143.1 CH954178 EDV51700.1 AE014296 AAN11811.1 AAN11812.2 BT023292 AAY55708.1 KK107413 EZA51158.1 CM000071 KRT02492.1 CCAG010007972 CH479183 EDW36173.1 EDY69147.1 QOIP01000005 RLU22850.1 KRT02493.1 CH933808 EDW10586.1 CH963894 EDW77160.1 OUUW01000004 SPP79394.1 CH916375 EDV98451.1 GL436923 EFN71338.1 GBYB01001800 JAG71567.1 KQ414648 KOC66187.1 GL446901 EFN87137.1 NNAY01000312 OXU29298.1 KQ435844 KOX71240.1 KQ981972 KYN31634.1 ADTU01002930 KQ979074 KYN22816.1 KQ976434 KYM87159.1 APGK01026367 APGK01026368 KB740627 ENN79940.1 DS235172 EEB12987.1 APGK01008292 APGK01008293 KB737656 ENN82984.1 LNIX01000006 OXA53201.1 MWRG01001773 PRD33677.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000037510 UP000075901 UP000075920 UP000069940 UP000249989 UP000076408 UP000075900 UP000008820 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000075884 UP000069272 UP000030765 UP000075880 UP000075881 UP000002320 UP000075885 UP000007266 UP000183832 UP000092553 UP000192223 UP000095300 UP000037069 UP000242457 UP000030742 UP000001292 UP000005203 UP000002282 UP000092443 UP000007801 UP000095301 UP000092460 UP000008792 UP000192221 UP000092445 UP000008711 UP000000803 UP000078200 UP000053097 UP000001819 UP000092444 UP000008744 UP000279307 UP000009192 UP000007798 UP000268350 UP000001070 UP000000311 UP000053825 UP000008237 UP000215335 UP000002358 UP000053105 UP000078541 UP000005205 UP000075902 UP000078492 UP000078540 UP000019118 UP000079169 UP000009046 UP000198287
UP000037510 UP000075901 UP000075920 UP000069940 UP000249989 UP000076408 UP000075900 UP000008820 UP000075882 UP000076407 UP000007062 UP000075840 UP000075886 UP000075884 UP000069272 UP000030765 UP000075880 UP000075881 UP000002320 UP000075885 UP000007266 UP000183832 UP000092553 UP000192223 UP000095300 UP000037069 UP000242457 UP000030742 UP000001292 UP000005203 UP000002282 UP000092443 UP000007801 UP000095301 UP000092460 UP000008792 UP000192221 UP000092445 UP000008711 UP000000803 UP000078200 UP000053097 UP000001819 UP000092444 UP000008744 UP000279307 UP000009192 UP000007798 UP000268350 UP000001070 UP000000311 UP000053825 UP000008237 UP000215335 UP000002358 UP000053105 UP000078541 UP000005205 UP000075902 UP000078492 UP000078540 UP000019118 UP000079169 UP000009046 UP000198287
Pfam
Interpro
IPR018499
Tetraspanin/Peripherin
+ More
IPR008952 Tetraspanin_EC2_sf
IPR000301 Tetraspanin
IPR008728 Elongator_complex_protein_4
IPR033828 GATase1_CTP_Synthase
IPR004468 CTP_synthase
IPR029062 Class_I_gatase-like
IPR017456 CTP_synthase_N
IPR017926 GATASE
IPR027417 P-loop_NTPase
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR034180 MCAD
IPR009075 AcylCo_DH/oxidase_C
IPR018503 Tetraspanin_CS
IPR008952 Tetraspanin_EC2_sf
IPR000301 Tetraspanin
IPR008728 Elongator_complex_protein_4
IPR033828 GATase1_CTP_Synthase
IPR004468 CTP_synthase
IPR029062 Class_I_gatase-like
IPR017456 CTP_synthase_N
IPR017926 GATASE
IPR027417 P-loop_NTPase
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR037069 AcylCoA_DH/ox_N_sf
IPR036250 AcylCo_DH-like_C
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009100 AcylCoA_DH/oxidase_NM_dom
IPR034180 MCAD
IPR009075 AcylCo_DH/oxidase_C
IPR018503 Tetraspanin_CS
SUPFAM
Gene 3D
ProteinModelPortal
H9IVG0
A0A2A4K4M5
A0A2H1VDC6
A0A194PD23
A0A3S2NZD4
A0A194RLJ0
+ More
A0A212FI16 A0A0L7LVL0 A0A182SEV8 A0A182W3P9 A0A023EQE6 A0A182YLZ4 A0A182RRD9 Q17D34 A0A182LN67 A0A182XAZ4 Q7QDT4 A0A182HX67 A0A1Q3FGF4 A0A182QPI8 A0A182NDI4 A0A182FM21 U5EX76 A0A084VTR7 A0A182IJ08 A0A182JWN9 A0A336MHF1 B0WVJ3 A0A182PUI8 D6W8H3 A0A1J1IIN1 A0A336KY22 A0A0M4E8N6 A0A0J9UKN3 A0A1W4XDW9 A0A1I8Q7V8 A0A1Y1KBZ6 A0A0L0C3W9 A0A2A3EKG1 U4UB82 A0A0A1WZW0 B4HH84 A0A088A4X2 B4PHW3 A0A0T6B8L0 A0A1A9XX71 B3MAN0 A0A1I8M1X7 A0A1B0AUW5 B4LNS3 A0A1W4UI57 A0A1A9Z0H7 B3NHH4 Q8IQK9 A0A1A9V2R0 Q4V3R4 A0A026W585 A0A0R3NNW6 A0A1B0FCD9 B4GI53 B5DZ48 A0A3L8DQX7 A0A0R3NTN9 B4KT74 B4MYH1 A0A3B0JAP4 B4JVI3 E2A5E6 A0A0C9R1H1 A0A0L7R5M4 E2BB46 A0A232FFR5 K7J7Z6 A0A0M8ZU68 A0A151JT85 A0A158NY22 A0A182UE64 A0A195ECI0 A0A151I588 N6UE05 A0A1S4EMK9 E0VHY1 A0A3Q0JH04 N6TR85 A0A226E8K7 A0A2P6L519
A0A212FI16 A0A0L7LVL0 A0A182SEV8 A0A182W3P9 A0A023EQE6 A0A182YLZ4 A0A182RRD9 Q17D34 A0A182LN67 A0A182XAZ4 Q7QDT4 A0A182HX67 A0A1Q3FGF4 A0A182QPI8 A0A182NDI4 A0A182FM21 U5EX76 A0A084VTR7 A0A182IJ08 A0A182JWN9 A0A336MHF1 B0WVJ3 A0A182PUI8 D6W8H3 A0A1J1IIN1 A0A336KY22 A0A0M4E8N6 A0A0J9UKN3 A0A1W4XDW9 A0A1I8Q7V8 A0A1Y1KBZ6 A0A0L0C3W9 A0A2A3EKG1 U4UB82 A0A0A1WZW0 B4HH84 A0A088A4X2 B4PHW3 A0A0T6B8L0 A0A1A9XX71 B3MAN0 A0A1I8M1X7 A0A1B0AUW5 B4LNS3 A0A1W4UI57 A0A1A9Z0H7 B3NHH4 Q8IQK9 A0A1A9V2R0 Q4V3R4 A0A026W585 A0A0R3NNW6 A0A1B0FCD9 B4GI53 B5DZ48 A0A3L8DQX7 A0A0R3NTN9 B4KT74 B4MYH1 A0A3B0JAP4 B4JVI3 E2A5E6 A0A0C9R1H1 A0A0L7R5M4 E2BB46 A0A232FFR5 K7J7Z6 A0A0M8ZU68 A0A151JT85 A0A158NY22 A0A182UE64 A0A195ECI0 A0A151I588 N6UE05 A0A1S4EMK9 E0VHY1 A0A3Q0JH04 N6TR85 A0A226E8K7 A0A2P6L519
Ontologies
GO
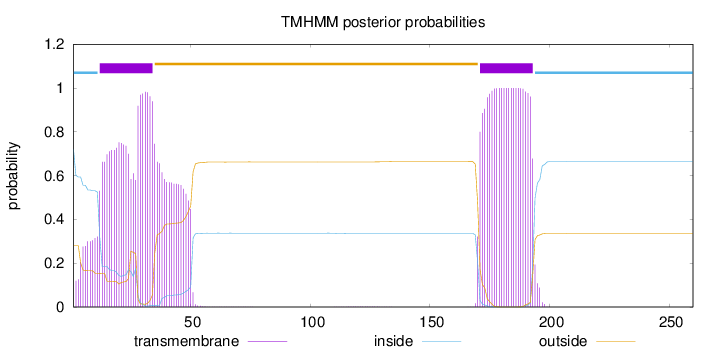
Topology
Subcellular location
Membrane
Length:
260
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
52.3581
Exp number, first 60 AAs:
29.45056
Total prob of N-in:
0.71998
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 170
TMhelix
171 - 193
inside
194 - 260
Population Genetic Test Statistics
Pi
198.579517
Theta
177.140755
Tajima's D
0.392082
CLR
0
CSRT
0.480025998700065
Interpretation
Uncertain
