Gene
KWMTBOMO07776 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001241
Annotation
heat_shock_protein_75_kDa?_mitochondrial-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.021
Sequence
CDS
ATGTTTGCAGTCCGTCCCAGAGTGGGAACATTCTATTTGAAAAGAATTTTATCCCGTTCAATGACTAATGTAATAATTGTAGCCCCGGGATGTGAAATGATAACCTCAAGACCAACGAATGTTTTCTTCGGATCCAAATTAAAAGAATTCCAACACTTTAGTACTGTAACTGAAAAACAAAGCAGTGAAAGCTTAAAAGGAAAGTCTGAAAAAAGAGAATTTCAAGCCGAAACGAGAATGTTGCTAGATATTGTAGCTCGATCTTTATATTCAGATAAAGAGGTATTTATTAGAGAGCTAATATCTAATGCAAGTGATGCTCTTGAAAAATTCAGATACCTGAGTGTAAGCTCAGTACCAGAGTCTGGAAACCTAAAAGAATTGGACAGACCACTTGAAATCAGATTAACAACTGACAAACAAAATCGTACAATTACATTCCAGGACTCTGGTATTGGTATGACGAAAGAAGAACTCACACAAAATTTAGGAACCATAGCACGATCTGGATCAAAGTCATTCATTGAAGAAATTAAAAAACAAGGTGCTGAACAAGCCAGCTCCATAATTGGGCAATTTGGTGTTGGATTCTATTCTGCATTTATGGTGGCTGACAAAATAGAAGTATTCACAAGAAGCAGTAAAGGCGATTCACCAGGATATCAATGGTCATCAGATGGATCTGGCACATATGAAATTCAAGAAGCTGATGGAGTTGAAATTGGCACAAAAATAGTTGTATATTTAAAAACAGACTGCAGAGAATTTGCAGATGATGATACAGTAAAAAGTATCATTAAGAAATACAGCAATTTTGTTAGCAGCCCTATTTTTTTGAACAACGACCAAGTTAATTCTATTAAGCCTTTATGGCTATTTGAACCGAAAGAAATAACTCAAGAGCAACATATTGAATTTTACAAATTCATCAGTAATTCATATGACAAACCTCGTTTCACACTTCACTACAAAACTGATGCGCCATTGAGTATAAGATCTATATTGTATGTACCAGAAGGGAAGCCTGGTCTCTTTGAGCTGTCACGTGATTCTGATGTAGGAGTGGCATTGTACTCCAGGAAGATATTGATTAAAAGCAAAGCAGAAAATATACTACCTAAATGGTTGCGATTTGTAAAAGGGGTTGTAGACTCTGAAGACATTCCGCTAAATCTCAGCAGAGAATTGTTACAAAACAGTGCGCTTATTACTAAACTGAGAACAGTCCTAACAAACCGGTTTCTGAAGTTTATGGTTGAAATGTCACAGAAAGATCCTTTGCAGTATGAGGCATTTTACAAAGACTACTCCATATTTCTGAAAGAGGGTATAGTTACAAGTCAAAGCCCATTGGAAAAGGAAGAAATTGCGAAATTATTACGATTTGAGTCATCGAAACTGAGTAATGGTCAAAGAACTTCCCTATCGGATTATAGTTCACGACAGAAAGATGATCAAAAGTCAATCTTTTATCTAGCAGCTCCTAGTCGAGAACTTGCAGAAACATCGCCATACTACGAATCACTTAAAAAACGCGACATCGAAGTATTGTTTTGCTATGAAATGTATGATGAGTTGGTGTTGTTAGAACTCAAAGAATTCGGTGGTCGAGCTTTAGTTTCAGTTGAAAACGACATGCAGAGAGATCCACTAGATAAATCGGAATCTATTATAGGCAGTGAGGGCCTGCAGCAATCTCAAGTGGATGAGTTAATCTCTTTTCTGAAAACGAACTTAACTGGGAAAGTATTTGATGTGCGCACAACGCAGAAGCTAGATTCTCATCCTTGCGTCATTGTCGTACCAGAAATGGCGGCTGCGAGACATTTCATTAGAACACAAGCTCAAAACCTCACTGAGGAGAACAGATTTGCACTTTTACGACCACAGCTAGAAATAAATGCTAAGCATCCAATTGTGAAAAAGTTACATAAACTGATGTCGACTGACAAGGAACTAGCGACCATGGTAGCTCAGCAGTTGTTCTCCAACGCCATGGTGACGGCTGGACTCGTCATGGATCCGAGAAATTTAATAACACATATAAATGATCTTCTAGTCAAGGTTTTAGAAAAACACTGA
Protein
MFAVRPRVGTFYLKRILSRSMTNVIIVAPGCEMITSRPTNVFFGSKLKEFQHFSTVTEKQSSESLKGKSEKREFQAETRMLLDIVARSLYSDKEVFIRELISNASDALEKFRYLSVSSVPESGNLKELDRPLEIRLTTDKQNRTITFQDSGIGMTKEELTQNLGTIARSGSKSFIEEIKKQGAEQASSIIGQFGVGFYSAFMVADKIEVFTRSSKGDSPGYQWSSDGSGTYEIQEADGVEIGTKIVVYLKTDCREFADDDTVKSIIKKYSNFVSSPIFLNNDQVNSIKPLWLFEPKEITQEQHIEFYKFISNSYDKPRFTLHYKTDAPLSIRSILYVPEGKPGLFELSRDSDVGVALYSRKILIKSKAENILPKWLRFVKGVVDSEDIPLNLSRELLQNSALITKLRTVLTNRFLKFMVEMSQKDPLQYEAFYKDYSIFLKEGIVTSQSPLEKEEIAKLLRFESSKLSNGQRTSLSDYSSRQKDDQKSIFYLAAPSRELAETSPYYESLKKRDIEVLFCYEMYDELVLLELKEFGGRALVSVENDMQRDPLDKSESIIGSEGLQQSQVDELISFLKTNLTGKVFDVRTTQKLDSHPCVIVVPEMAAARHFIRTQAQNLTEENRFALLRPQLEINAKHPIVKKLHKLMSTDKELATMVAQQLFSNAMVTAGLVMDPRNLITHINDLLVKVLEKH
Summary
Uniprot
H9U397
H9IVG1
A0A3S2L912
A0A194PDW9
A0A194RGA3
A0A2A4K4C1
+ More
A0A212FHZ0 A0A2H1V6S3 Q17PR3 A0A182G262 B0WV61 A0A182YS12 A0A2M4ABH9 A0A2M4CN11 A0A182FKH2 A0A182VM43 A0A2M4ABC0 A0A182RLU5 A0A2M4BFI2 A0A2M3Z2D6 A0A2M4BFM4 A0A182THM4 A0A1Q3FFE7 A0A182K6P5 A0A182P7K5 A0A182WE76 A0A182I612 A0A182WXZ2 A0A1S4H5I6 A0A182KUZ8 Q7QE24 A0A182JJ05 A0A2M4CN64 A0A182M1Y9 W5JQ40 A0A182QXL1 A0A084WUD5 A0A182NPA8 A0A0K8U9W6 A0A034VAK1 A0A034V672 U5ETP0 A0A336LXU5 A0A0A1XBM5 A0A1I8NUU1 W8C0J6 A0A1I8M2Y4 A0A2M4BHD5 T1PG34 A0A1Y1LQC6 A0A0L0BW03 A0A1L8DUA6 A0A2J7PFN7 A0A2M3Z3H6 A0A1B0GI32 A0A1B6DQJ3 Q8IHC2 B4MJZ2 E0VHX6 A0A1B6FTU8 B4QD97 A0A1B6JSP9 B4LIM6 B4HPV6 Q7KNF3 B4P0Y2 A1Z6L9 A0A0C9RMR0 B3MJ48 N6U4W9 A0A1B6L5M4 B3NB53 A0A1W4VEQ3 A0A0P5ZBB1 A0A0N8AWJ0 A0A0P4ZII4 Q28WX4 A0A0P5VGQ3 A0A0P5ZB67 B4JVQ8 A0A3B0J2K2 B4KUH3 A0A0P5U176 A0A0P4ZZC6 A0A0P5V322 A0A0P5B7K9 A0A1J1IRI1 A0A023EV66 A0A0M4EBM2 A0A1B0D5R4 E9GIU3 A0A232FFT5 A0A1W4X5B4 A0A1D2N1P9 A0A386GY33 A0A026WHX1 E2AER8 F4X0W3 A0A151WGT0 A0A154PDV5
A0A212FHZ0 A0A2H1V6S3 Q17PR3 A0A182G262 B0WV61 A0A182YS12 A0A2M4ABH9 A0A2M4CN11 A0A182FKH2 A0A182VM43 A0A2M4ABC0 A0A182RLU5 A0A2M4BFI2 A0A2M3Z2D6 A0A2M4BFM4 A0A182THM4 A0A1Q3FFE7 A0A182K6P5 A0A182P7K5 A0A182WE76 A0A182I612 A0A182WXZ2 A0A1S4H5I6 A0A182KUZ8 Q7QE24 A0A182JJ05 A0A2M4CN64 A0A182M1Y9 W5JQ40 A0A182QXL1 A0A084WUD5 A0A182NPA8 A0A0K8U9W6 A0A034VAK1 A0A034V672 U5ETP0 A0A336LXU5 A0A0A1XBM5 A0A1I8NUU1 W8C0J6 A0A1I8M2Y4 A0A2M4BHD5 T1PG34 A0A1Y1LQC6 A0A0L0BW03 A0A1L8DUA6 A0A2J7PFN7 A0A2M3Z3H6 A0A1B0GI32 A0A1B6DQJ3 Q8IHC2 B4MJZ2 E0VHX6 A0A1B6FTU8 B4QD97 A0A1B6JSP9 B4LIM6 B4HPV6 Q7KNF3 B4P0Y2 A1Z6L9 A0A0C9RMR0 B3MJ48 N6U4W9 A0A1B6L5M4 B3NB53 A0A1W4VEQ3 A0A0P5ZBB1 A0A0N8AWJ0 A0A0P4ZII4 Q28WX4 A0A0P5VGQ3 A0A0P5ZB67 B4JVQ8 A0A3B0J2K2 B4KUH3 A0A0P5U176 A0A0P4ZZC6 A0A0P5V322 A0A0P5B7K9 A0A1J1IRI1 A0A023EV66 A0A0M4EBM2 A0A1B0D5R4 E9GIU3 A0A232FFT5 A0A1W4X5B4 A0A1D2N1P9 A0A386GY33 A0A026WHX1 E2AER8 F4X0W3 A0A151WGT0 A0A154PDV5
Pubmed
19121390
26354079
22118469
17510324
26483478
25244985
+ More
12364791 20966253 20920257 23761445 24438588 25348373 25830018 24495485 25315136 28004739 26108605 17994087 20566863 22936249 10652318 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 15632085 24945155 21292972 28648823 27289101 30226893 24508170 30249741 20798317 21719571
12364791 20966253 20920257 23761445 24438588 25348373 25830018 24495485 25315136 28004739 26108605 17994087 20566863 22936249 10652318 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 15632085 24945155 21292972 28648823 27289101 30226893 24508170 30249741 20798317 21719571
EMBL
JQ313137
AFG30049.1
BABH01001041
RSAL01000084
RVE48415.1
KQ459606
+ More
KPI91218.1 KQ460205 KPJ16848.1 NWSH01000189 PCG78603.1 AGBW02008449 OWR53364.1 ODYU01000982 SOQ36555.1 CH477190 EAT48688.1 JXUM01039146 KQ561192 KXJ79416.1 DS232119 EDS35400.1 GGFK01004751 MBW38072.1 GGFL01002538 MBW66716.1 GGFK01004763 MBW38084.1 GGFJ01002651 MBW51792.1 GGFM01001941 MBW22692.1 GGFJ01002652 MBW51793.1 GFDL01008775 JAV26270.1 APCN01003545 AAAB01008848 EAA07036.3 GGFL01002537 MBW66715.1 AXCM01001139 ADMH02000484 ETN66256.1 AXCN02001819 ATLV01027079 KE525423 KFB53829.1 GDHF01028852 JAI23462.1 GAKP01020162 JAC38790.1 GAKP01020161 JAC38791.1 GANO01002691 JAB57180.1 UFQT01000292 SSX22886.1 GBXI01005946 JAD08346.1 GAMC01003982 JAC02574.1 GGFJ01003325 MBW52466.1 KA647777 AFP62406.1 GEZM01049781 JAV75849.1 JRES01001352 KNC23389.1 GFDF01004098 JAV09986.1 NEVH01025655 PNF15137.1 GGFM01002247 MBW22998.1 AJWK01011218 GEDC01009358 JAS27940.1 BT001316 AAN71071.1 CH963846 EDW72431.1 DS235172 EEB12982.1 GECZ01016139 JAS53630.1 CM000362 CM002911 EDX05884.1 KMY91749.1 GECU01005487 JAT02220.1 CH940648 EDW60396.1 CH480816 EDW46625.1 AF115775 AAD29307.2 CM000157 EDW89056.1 AE013599 BT150318 AAF57362.1 AGB93259.1 AGW25600.1 GBYB01008276 JAG78043.1 CH902619 EDV38142.1 APGK01039339 KB740970 KB632388 ENN76640.1 ERL94369.1 GEBQ01020959 JAT19018.1 CH954177 EDV59818.2 GDIP01059044 JAM44671.1 GDIQ01235939 JAK15786.1 GDIP01225523 JAI97878.1 CM000071 EAL26542.1 GDIP01101880 JAM01835.1 GDIP01047156 LRGB01002993 JAM56559.1 KZS05193.1 CH916375 EDV98046.1 OUUW01000001 SPP73342.1 CH933808 EDW10760.1 GDIP01119584 JAL84130.1 GDIP01206116 JAJ17286.1 GDIP01104835 JAL98879.1 GDIP01189222 JAJ34180.1 CVRI01000056 CRL01686.1 GAPW01000712 JAC12886.1 CP012524 ALC42684.1 AJVK01025536 GL732547 EFX80327.1 NNAY01000312 OXU29299.1 LJIJ01000294 ODM99213.1 MH490979 AYD37992.1 KK107214 QOIP01000005 EZA55266.1 RLU22479.1 GL438929 EFN68064.1 GL888503 EGI59905.1 KQ983152 KYQ47015.1 KQ434886 KZC10076.1
KPI91218.1 KQ460205 KPJ16848.1 NWSH01000189 PCG78603.1 AGBW02008449 OWR53364.1 ODYU01000982 SOQ36555.1 CH477190 EAT48688.1 JXUM01039146 KQ561192 KXJ79416.1 DS232119 EDS35400.1 GGFK01004751 MBW38072.1 GGFL01002538 MBW66716.1 GGFK01004763 MBW38084.1 GGFJ01002651 MBW51792.1 GGFM01001941 MBW22692.1 GGFJ01002652 MBW51793.1 GFDL01008775 JAV26270.1 APCN01003545 AAAB01008848 EAA07036.3 GGFL01002537 MBW66715.1 AXCM01001139 ADMH02000484 ETN66256.1 AXCN02001819 ATLV01027079 KE525423 KFB53829.1 GDHF01028852 JAI23462.1 GAKP01020162 JAC38790.1 GAKP01020161 JAC38791.1 GANO01002691 JAB57180.1 UFQT01000292 SSX22886.1 GBXI01005946 JAD08346.1 GAMC01003982 JAC02574.1 GGFJ01003325 MBW52466.1 KA647777 AFP62406.1 GEZM01049781 JAV75849.1 JRES01001352 KNC23389.1 GFDF01004098 JAV09986.1 NEVH01025655 PNF15137.1 GGFM01002247 MBW22998.1 AJWK01011218 GEDC01009358 JAS27940.1 BT001316 AAN71071.1 CH963846 EDW72431.1 DS235172 EEB12982.1 GECZ01016139 JAS53630.1 CM000362 CM002911 EDX05884.1 KMY91749.1 GECU01005487 JAT02220.1 CH940648 EDW60396.1 CH480816 EDW46625.1 AF115775 AAD29307.2 CM000157 EDW89056.1 AE013599 BT150318 AAF57362.1 AGB93259.1 AGW25600.1 GBYB01008276 JAG78043.1 CH902619 EDV38142.1 APGK01039339 KB740970 KB632388 ENN76640.1 ERL94369.1 GEBQ01020959 JAT19018.1 CH954177 EDV59818.2 GDIP01059044 JAM44671.1 GDIQ01235939 JAK15786.1 GDIP01225523 JAI97878.1 CM000071 EAL26542.1 GDIP01101880 JAM01835.1 GDIP01047156 LRGB01002993 JAM56559.1 KZS05193.1 CH916375 EDV98046.1 OUUW01000001 SPP73342.1 CH933808 EDW10760.1 GDIP01119584 JAL84130.1 GDIP01206116 JAJ17286.1 GDIP01104835 JAL98879.1 GDIP01189222 JAJ34180.1 CVRI01000056 CRL01686.1 GAPW01000712 JAC12886.1 CP012524 ALC42684.1 AJVK01025536 GL732547 EFX80327.1 NNAY01000312 OXU29299.1 LJIJ01000294 ODM99213.1 MH490979 AYD37992.1 KK107214 QOIP01000005 EZA55266.1 RLU22479.1 GL438929 EFN68064.1 GL888503 EGI59905.1 KQ983152 KYQ47015.1 KQ434886 KZC10076.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000218220
UP000007151
+ More
UP000008820 UP000069940 UP000249989 UP000002320 UP000076408 UP000069272 UP000075903 UP000075900 UP000075902 UP000075881 UP000075885 UP000075920 UP000075840 UP000076407 UP000075882 UP000007062 UP000075880 UP000075883 UP000000673 UP000075886 UP000030765 UP000075884 UP000095300 UP000095301 UP000037069 UP000235965 UP000092461 UP000007798 UP000009046 UP000000304 UP000008792 UP000001292 UP000002282 UP000000803 UP000007801 UP000019118 UP000030742 UP000008711 UP000192221 UP000001819 UP000076858 UP000001070 UP000268350 UP000009192 UP000183832 UP000092553 UP000092462 UP000000305 UP000215335 UP000192223 UP000094527 UP000053097 UP000279307 UP000000311 UP000007755 UP000075809 UP000076502
UP000008820 UP000069940 UP000249989 UP000002320 UP000076408 UP000069272 UP000075903 UP000075900 UP000075902 UP000075881 UP000075885 UP000075920 UP000075840 UP000076407 UP000075882 UP000007062 UP000075880 UP000075883 UP000000673 UP000075886 UP000030765 UP000075884 UP000095300 UP000095301 UP000037069 UP000235965 UP000092461 UP000007798 UP000009046 UP000000304 UP000008792 UP000001292 UP000002282 UP000000803 UP000007801 UP000019118 UP000030742 UP000008711 UP000192221 UP000001819 UP000076858 UP000001070 UP000268350 UP000009192 UP000183832 UP000092553 UP000092462 UP000000305 UP000215335 UP000192223 UP000094527 UP000053097 UP000279307 UP000000311 UP000007755 UP000075809 UP000076502
Pfam
PF00183 HSP90
Interpro
Gene 3D
ProteinModelPortal
H9U397
H9IVG1
A0A3S2L912
A0A194PDW9
A0A194RGA3
A0A2A4K4C1
+ More
A0A212FHZ0 A0A2H1V6S3 Q17PR3 A0A182G262 B0WV61 A0A182YS12 A0A2M4ABH9 A0A2M4CN11 A0A182FKH2 A0A182VM43 A0A2M4ABC0 A0A182RLU5 A0A2M4BFI2 A0A2M3Z2D6 A0A2M4BFM4 A0A182THM4 A0A1Q3FFE7 A0A182K6P5 A0A182P7K5 A0A182WE76 A0A182I612 A0A182WXZ2 A0A1S4H5I6 A0A182KUZ8 Q7QE24 A0A182JJ05 A0A2M4CN64 A0A182M1Y9 W5JQ40 A0A182QXL1 A0A084WUD5 A0A182NPA8 A0A0K8U9W6 A0A034VAK1 A0A034V672 U5ETP0 A0A336LXU5 A0A0A1XBM5 A0A1I8NUU1 W8C0J6 A0A1I8M2Y4 A0A2M4BHD5 T1PG34 A0A1Y1LQC6 A0A0L0BW03 A0A1L8DUA6 A0A2J7PFN7 A0A2M3Z3H6 A0A1B0GI32 A0A1B6DQJ3 Q8IHC2 B4MJZ2 E0VHX6 A0A1B6FTU8 B4QD97 A0A1B6JSP9 B4LIM6 B4HPV6 Q7KNF3 B4P0Y2 A1Z6L9 A0A0C9RMR0 B3MJ48 N6U4W9 A0A1B6L5M4 B3NB53 A0A1W4VEQ3 A0A0P5ZBB1 A0A0N8AWJ0 A0A0P4ZII4 Q28WX4 A0A0P5VGQ3 A0A0P5ZB67 B4JVQ8 A0A3B0J2K2 B4KUH3 A0A0P5U176 A0A0P4ZZC6 A0A0P5V322 A0A0P5B7K9 A0A1J1IRI1 A0A023EV66 A0A0M4EBM2 A0A1B0D5R4 E9GIU3 A0A232FFT5 A0A1W4X5B4 A0A1D2N1P9 A0A386GY33 A0A026WHX1 E2AER8 F4X0W3 A0A151WGT0 A0A154PDV5
A0A212FHZ0 A0A2H1V6S3 Q17PR3 A0A182G262 B0WV61 A0A182YS12 A0A2M4ABH9 A0A2M4CN11 A0A182FKH2 A0A182VM43 A0A2M4ABC0 A0A182RLU5 A0A2M4BFI2 A0A2M3Z2D6 A0A2M4BFM4 A0A182THM4 A0A1Q3FFE7 A0A182K6P5 A0A182P7K5 A0A182WE76 A0A182I612 A0A182WXZ2 A0A1S4H5I6 A0A182KUZ8 Q7QE24 A0A182JJ05 A0A2M4CN64 A0A182M1Y9 W5JQ40 A0A182QXL1 A0A084WUD5 A0A182NPA8 A0A0K8U9W6 A0A034VAK1 A0A034V672 U5ETP0 A0A336LXU5 A0A0A1XBM5 A0A1I8NUU1 W8C0J6 A0A1I8M2Y4 A0A2M4BHD5 T1PG34 A0A1Y1LQC6 A0A0L0BW03 A0A1L8DUA6 A0A2J7PFN7 A0A2M3Z3H6 A0A1B0GI32 A0A1B6DQJ3 Q8IHC2 B4MJZ2 E0VHX6 A0A1B6FTU8 B4QD97 A0A1B6JSP9 B4LIM6 B4HPV6 Q7KNF3 B4P0Y2 A1Z6L9 A0A0C9RMR0 B3MJ48 N6U4W9 A0A1B6L5M4 B3NB53 A0A1W4VEQ3 A0A0P5ZBB1 A0A0N8AWJ0 A0A0P4ZII4 Q28WX4 A0A0P5VGQ3 A0A0P5ZB67 B4JVQ8 A0A3B0J2K2 B4KUH3 A0A0P5U176 A0A0P4ZZC6 A0A0P5V322 A0A0P5B7K9 A0A1J1IRI1 A0A023EV66 A0A0M4EBM2 A0A1B0D5R4 E9GIU3 A0A232FFT5 A0A1W4X5B4 A0A1D2N1P9 A0A386GY33 A0A026WHX1 E2AER8 F4X0W3 A0A151WGT0 A0A154PDV5
PDB
4IYN
E-value=0,
Score=1827
Ontologies
GO
PANTHER
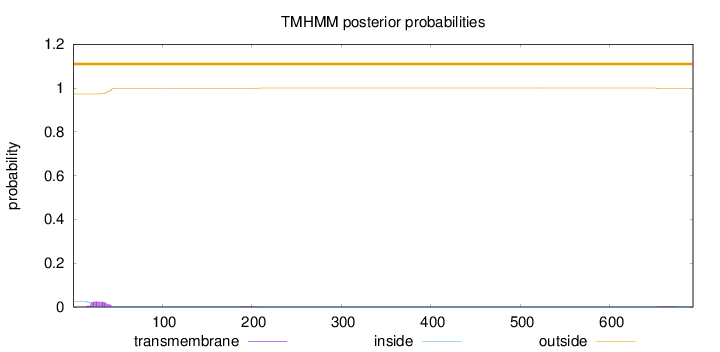
Topology
Length:
693
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.60605
Exp number, first 60 AAs:
0.50626
Total prob of N-in:
0.02670
outside
1 - 693
Population Genetic Test Statistics
Pi
233.359365
Theta
19.699861
Tajima's D
1.024303
CLR
0
CSRT
0.671716414179291
Interpretation
Uncertain
