Pre Gene Modal
BGIBMGA001242
Annotation
PREDICTED:_neurotactin_isoform_X2_[Bombyx_mori]
Full name
Neurotactin
Location in the cell
Cytoplasmic Reliability : 2.001
Sequence
CDS
ATGAGTACAACTGAAATTAATAAAATAGAGTTTGAGGATGAGAAGACGTTGGATAAAAAGGAAATTGAAGCCGAAGAGAAAGAAGTTATGCTAAAAGCCGAAAAGGAAGCGATTGATGATTTTGCGGAACTGAGAGCTGAAGCAGAAGGACCTGTAAAGGCAGCTCTTTCTGGTGATGTTACGGAACAAGGACGCGAAGTCAAGCCGAAATATATACCTATCGGGGCCATCAAAATGCCGGGATTTTTCACGAAAAGTTCTGAAAAATCGAAGGATGAAGAAGAAATTCAGAAAGAAACAGATATCTGCAAGACTGATGAAACTTCGAAACCAAAAGATGAACACAAAGAACAGCAAAAAACCAAAAGAAGTATATTTAATATCAGATATCCAAATTTTTTCAAAAAACGTTCGCCGAGTACTGCCGAGGCCACACTAGCTTCAATGGAAACTTTGGATGATAAAATGGATACATCTAATGATGGATTGGAAAATGTAAAACTGGAACCTATGGATCCTGAAGATGGCAAAGTGGCCACTAAACTTCCCTTGAAAGAAAGAATTCGACAAAAGAAATTTATCATTGACGACATTGTGGTTTGCATTGTGGCGCTCTTAGTGGCCATGTCGGTGATCATTGGAAGCTTGATTGGAGCTTGTGTCGGACCGCCAGTTGAACGACCTTTGCGTTTGGGACGTTATATCACCGCGTCTACCGGCTGCGGACCCGTGGAAGGAATATTGGATGAAGGTGTTTACAAGTTCTACAGCATTCCATATGCAGTTCCTCCAATAAAAGAGAAACGGTTTACCTACGCACAACCATTAAACAATTTATCGCTATGTTGGAATGAAACTTATCAATGCCACGTGCCTGGGCCACTCTGCGTTCAGTTTCTGGAGAACGGCACTATTACTGGCGAAGAAGACTGCCTCACGCTCGACATTATGACGCCTCACGTCAGGTACGACTCACCGTTGCCCGTAGTTGTACTCATCGGGGCTAACACACTTGCTGGCGGAATAAGCCCAGCTCAACCATCAGCTTTGTATGCTCGGACCGAGGAAGTTATTTTTGTGCGCCCCAACTTTAGACTAGGATCTTTCGGTTTTTTATCTCTCAATATACTATCCAAGTCAAAATACCCGCATACCTCTGGCAACTACGCTCTTAGCGATTTGCTGGCTGCACTTCGTTGGATCCATTTAAACATTAATCATTTTGGAGGAGATCCAGAAATGGTAACCTTACTTGGTCATCGAGCTGGAGCGACATTAACTGCCGCTTTGACGACAGTCTCCGGAGCGCACAAGCTGTACACCAGAGTTTGGCTGTCTTCACCTTCTGTAATATTTCCCGGAGAAGTTCTAGAGCAGTCTCAGAAGAACAACGAACAATTCAGGCAGATGAGTAAATGTAATGATGCGGATTGTCTGAGACGTGTGTCCAAAGAAGAGCTGTTGGCGCTCACGCCTGATACCTGGTTCGGAAATAATTTTGAGACGTTGCCCCGGACTGACGAAATTCAACGGTCTTGGTTAGTCTTGGACGGAGAGATGTTACGTATTCACGCGTACGAAAGTTGGCTCGATCAGAAGACTGCAAAACAATCTGGTAAAGTAAAAGTTTTCAAACCGATGGTCTTCACCACGACCCTGCATTCCGGCCACTCAGTTACTCTGAAAAACAAACACGTCAAGTGGACACCGGAGATCGTAGAAAAAATCGTTAACGACAGCGTCATCGGGGAGAAGAACTTGACCGCCGCCGTATTCACACATTTTAATAAGACTTACGAGGGTCTAGTAGAACTGATTTCGTCTGTCCGAACTCTGTGCCCGCTGGTGAGCCTGGCGCGGCTGCGGCTGTCGGTCCCGCTGTACGTAGTGCGGGGCGCGAGGCGGGGGGCGGAGGGGTCGCCTGGGCTGGCCGACGTCAACGCGGACGTCAAAGCAATTCTCGGTACTTTTGATTCCGAGATGCCCGAACAGCGTAGATTCATGGCAGTCATCCAACACCTGTTCTACCATTACGTCGGGCACGGCAAGCTGTCCGGTCCCGAGCCGGGCCTCGTTGCGATTGGCCAGGACCTGCTTCCTCAGCCCAGCCTTCCCGCGTGCGATCTACTCATCCGAGAGGACATCGTCCCGCGATACGCTCACGTCGATTAG
Protein
MSTTEINKIEFEDEKTLDKKEIEAEEKEVMLKAEKEAIDDFAELRAEAEGPVKAALSGDVTEQGREVKPKYIPIGAIKMPGFFTKSSEKSKDEEEIQKETDICKTDETSKPKDEHKEQQKTKRSIFNIRYPNFFKKRSPSTAEATLASMETLDDKMDTSNDGLENVKLEPMDPEDGKVATKLPLKERIRQKKFIIDDIVVCIVALLVAMSVIIGSLIGACVGPPVERPLRLGRYITASTGCGPVEGILDEGVYKFYSIPYAVPPIKEKRFTYAQPLNNLSLCWNETYQCHVPGPLCVQFLENGTITGEEDCLTLDIMTPHVRYDSPLPVVVLIGANTLAGGISPAQPSALYARTEEVIFVRPNFRLGSFGFLSLNILSKSKYPHTSGNYALSDLLAALRWIHLNINHFGGDPEMVTLLGHRAGATLTAALTTVSGAHKLYTRVWLSSPSVIFPGEVLEQSQKNNEQFRQMSKCNDADCLRRVSKEELLALTPDTWFGNNFETLPRTDEIQRSWLVLDGEMLRIHAYESWLDQKTAKQSGKVKVFKPMVFTTTLHSGHSVTLKNKHVKWTPEIVEKIVNDSVIGEKNLTAAVFTHFNKTYEGLVELISSVRTLCPLVSLARLRLSVPLYVVRGARRGAEGSPGLADVNADVKAILGTFDSEMPEQRRFMAVIQHLFYHYVGHGKLSGPEPGLVAIGQDLLPQPSLPACDLLIREDIVPRYAHVD
Summary
Description
May mediate or modulate cell adhesion between embryonic cells during development.
Similarity
In the C-terminal section; belongs to the type-B carboxylesterase/lipase family.
Keywords
Cell adhesion
Complete proteome
Disulfide bond
Glycoprotein
Membrane
Phosphoprotein
Reference proteome
Signal-anchor
Transmembrane
Transmembrane helix
Feature
chain Neurotactin
Uniprot
H9IVG2
A0A2A4K3X3
A0A0U2LCP7
A0A3S2NTU0
A0A2H1V6Z2
A0A194PDA0
+ More
A0A1U9X1Y1 A0A0L7LDS9 A0A194RLI5 B0WNN8 A0A0L0BPS0 Q16WF1 W8C4D2 A0A182GAQ9 A0A182GJ52 A0A0K8UZL1 A0A068F7V0 A0A034VJG4 A0A0A1XRB1 A0A1A9XQZ3 A0A1B0C2N7 A0A1A9ZLW6 A0A1A9W4Y7 B4LG54 A0A1A9UP19 D2A2B8 B4N3X7 A0A182J1I1 A0A182K8Z1 A0A1I8M356 Q29DT7 A0A336L831 A0A182LJ43 A0A084WK64 A0A336LFM2 Q7Q6V8 A0A182VJS9 A0A182MVK1 A0A3B0JZQ0 A0A182FC87 A0A1I8PJ28 A0A182NGW2 A0A336M919 A0A182TE28 A0A182WII3 A0A182Y1A1 A0A182QGF7 A0A0P8XV10 A0A182RF94 B4KXZ6 A0A182SDN2 A0A182PHM2 W5JIH3 A0A1B0G7J2 B4IXF7 B3M4E3 A0A182XMX1 B4HK63 B4PKA2 A0A1W4WEL0 A0A0J9RWN7 M9MS15 P23654 B3NDH9 A0A0M4EFV3 A0A1W4UGW0 A0A154PKJ7 A0A0L7R8Y8 A0A310SCB4 A0A087ZXD6 A0A2A3EDS0 A0A3L8E242 A0A0J7KAZ7 A0A195FVE2 A0A158NW65 A0A195B871 F4WNG2 A0A195ED01 A0A151WVD4 A0A1B6MHX5 A0A0T6AYL4 A0A0C9RU64 A0A2J7R8H1 A0A1J1IXZ2 A0A026WLA0 K7IZ24 A0A067RH81 E2BI28 A0A232F7E5 A0A0N0U7U2 E0VCD0 A0A2P8Y5J5 A0A151IM92
A0A1U9X1Y1 A0A0L7LDS9 A0A194RLI5 B0WNN8 A0A0L0BPS0 Q16WF1 W8C4D2 A0A182GAQ9 A0A182GJ52 A0A0K8UZL1 A0A068F7V0 A0A034VJG4 A0A0A1XRB1 A0A1A9XQZ3 A0A1B0C2N7 A0A1A9ZLW6 A0A1A9W4Y7 B4LG54 A0A1A9UP19 D2A2B8 B4N3X7 A0A182J1I1 A0A182K8Z1 A0A1I8M356 Q29DT7 A0A336L831 A0A182LJ43 A0A084WK64 A0A336LFM2 Q7Q6V8 A0A182VJS9 A0A182MVK1 A0A3B0JZQ0 A0A182FC87 A0A1I8PJ28 A0A182NGW2 A0A336M919 A0A182TE28 A0A182WII3 A0A182Y1A1 A0A182QGF7 A0A0P8XV10 A0A182RF94 B4KXZ6 A0A182SDN2 A0A182PHM2 W5JIH3 A0A1B0G7J2 B4IXF7 B3M4E3 A0A182XMX1 B4HK63 B4PKA2 A0A1W4WEL0 A0A0J9RWN7 M9MS15 P23654 B3NDH9 A0A0M4EFV3 A0A1W4UGW0 A0A154PKJ7 A0A0L7R8Y8 A0A310SCB4 A0A087ZXD6 A0A2A3EDS0 A0A3L8E242 A0A0J7KAZ7 A0A195FVE2 A0A158NW65 A0A195B871 F4WNG2 A0A195ED01 A0A151WVD4 A0A1B6MHX5 A0A0T6AYL4 A0A0C9RU64 A0A2J7R8H1 A0A1J1IXZ2 A0A026WLA0 K7IZ24 A0A067RH81 E2BI28 A0A232F7E5 A0A0N0U7U2 E0VCD0 A0A2P8Y5J5 A0A151IM92
Pubmed
19121390
26749290
26354079
26227816
26108605
17510324
+ More
24495485 26483478 25880816 25348373 25830018 17994087 18057021 18362917 19820115 25315136 15632085 20966253 24438588 12364791 14747013 17210077 25244985 20920257 23761445 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2100266 2120047 10731138 2120048 18327897 30249741 21347285 21719571 24508170 20075255 24845553 20798317 28648823 20566863 29403074
24495485 26483478 25880816 25348373 25830018 17994087 18057021 18362917 19820115 25315136 15632085 20966253 24438588 12364791 14747013 17210077 25244985 20920257 23761445 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 2100266 2120047 10731138 2120048 18327897 30249741 21347285 21719571 24508170 20075255 24845553 20798317 28648823 20566863 29403074
EMBL
BABH01001041
NWSH01000189
PCG78604.1
KP308208
ALH43426.1
RSAL01000084
+ More
RVE48414.1 ODYU01000982 SOQ36556.1 KQ459606 KPI91217.1 KY021879 AQY62768.1 JTDY01001519 KOB73668.1 KQ460205 KPJ16846.1 DS232013 EDS31874.1 JRES01001646 KNC21219.1 CH477568 EAT38911.1 GAMC01002372 JAC04184.1 JXUM01051274 JXUM01051275 KQ561682 KXJ77811.1 JXUM01012574 KQ560357 KXJ82854.1 GDHF01030899 GDHF01020574 GDHF01002575 JAI21415.1 JAI31740.1 JAI49739.1 KJ702193 AID61347.1 GAKP01016318 JAC42634.1 GBXI01009547 GBXI01000780 JAD04745.1 JAD13512.1 JXJN01024608 CH940647 EDW69362.1 KRF84338.1 KRF84339.1 KQ971338 EFA02046.2 CH964095 EDW79332.1 CH379070 EAL30327.1 UFQS01001976 UFQT01001976 SSX12833.1 SSX32275.1 ATLV01024095 KE525349 KFB50608.1 SSX12832.1 SSX32274.1 AAAB01008960 EAA11248.4 AXCM01005329 OUUW01000012 SPP87555.1 UFQT01000574 SSX25409.1 AXCN02000363 CH902618 KPU78575.1 CH933809 EDW18698.1 KRG06320.1 ADMH02001299 ETN63093.1 CCAG010022573 CCAG010022574 CCAG010022575 CCAG010022576 CCAG010022577 CCAG010022578 CH916366 EDV96394.1 EDV40437.1 KPU78576.1 CH480815 EDW41804.1 CM000159 EDW94800.1 CM002912 KMZ00104.1 AE014296 ADV37557.1 X54999 X53837 AF132188 CH954178 EDV52041.1 CP012525 ALC44908.1 KQ434931 KZC11828.1 KQ414631 KOC67241.1 KQ763581 OAD54934.1 KZ288288 PBC29316.1 QOIP01000001 RLU26523.1 LBMM01010484 KMQ87431.1 KQ981215 KYN44610.1 ADTU01027829 ADTU01027830 KQ976565 KYM80445.1 GL888237 EGI64155.1 KQ979074 KYN22697.1 KQ982706 KYQ51834.1 GEBQ01004461 JAT35516.1 LJIG01022487 KRT80320.1 GBYB01012270 GBYB01012433 GBYB01012512 JAG82037.1 JAG82200.1 JAG82279.1 NEVH01006721 PNF37125.1 CVRI01000063 CRL04436.1 KK107154 EZA56832.1 KK852637 KDR19688.1 GL448433 EFN84654.1 NNAY01000773 OXU26595.1 KQ435689 KOX81251.1 DS235053 EEB11036.1 PYGN01000901 PSN39532.1 KQ977063 KYN06008.1
RVE48414.1 ODYU01000982 SOQ36556.1 KQ459606 KPI91217.1 KY021879 AQY62768.1 JTDY01001519 KOB73668.1 KQ460205 KPJ16846.1 DS232013 EDS31874.1 JRES01001646 KNC21219.1 CH477568 EAT38911.1 GAMC01002372 JAC04184.1 JXUM01051274 JXUM01051275 KQ561682 KXJ77811.1 JXUM01012574 KQ560357 KXJ82854.1 GDHF01030899 GDHF01020574 GDHF01002575 JAI21415.1 JAI31740.1 JAI49739.1 KJ702193 AID61347.1 GAKP01016318 JAC42634.1 GBXI01009547 GBXI01000780 JAD04745.1 JAD13512.1 JXJN01024608 CH940647 EDW69362.1 KRF84338.1 KRF84339.1 KQ971338 EFA02046.2 CH964095 EDW79332.1 CH379070 EAL30327.1 UFQS01001976 UFQT01001976 SSX12833.1 SSX32275.1 ATLV01024095 KE525349 KFB50608.1 SSX12832.1 SSX32274.1 AAAB01008960 EAA11248.4 AXCM01005329 OUUW01000012 SPP87555.1 UFQT01000574 SSX25409.1 AXCN02000363 CH902618 KPU78575.1 CH933809 EDW18698.1 KRG06320.1 ADMH02001299 ETN63093.1 CCAG010022573 CCAG010022574 CCAG010022575 CCAG010022576 CCAG010022577 CCAG010022578 CH916366 EDV96394.1 EDV40437.1 KPU78576.1 CH480815 EDW41804.1 CM000159 EDW94800.1 CM002912 KMZ00104.1 AE014296 ADV37557.1 X54999 X53837 AF132188 CH954178 EDV52041.1 CP012525 ALC44908.1 KQ434931 KZC11828.1 KQ414631 KOC67241.1 KQ763581 OAD54934.1 KZ288288 PBC29316.1 QOIP01000001 RLU26523.1 LBMM01010484 KMQ87431.1 KQ981215 KYN44610.1 ADTU01027829 ADTU01027830 KQ976565 KYM80445.1 GL888237 EGI64155.1 KQ979074 KYN22697.1 KQ982706 KYQ51834.1 GEBQ01004461 JAT35516.1 LJIG01022487 KRT80320.1 GBYB01012270 GBYB01012433 GBYB01012512 JAG82037.1 JAG82200.1 JAG82279.1 NEVH01006721 PNF37125.1 CVRI01000063 CRL04436.1 KK107154 EZA56832.1 KK852637 KDR19688.1 GL448433 EFN84654.1 NNAY01000773 OXU26595.1 KQ435689 KOX81251.1 DS235053 EEB11036.1 PYGN01000901 PSN39532.1 KQ977063 KYN06008.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000037510
UP000053240
+ More
UP000002320 UP000037069 UP000008820 UP000069940 UP000249989 UP000092443 UP000092460 UP000092445 UP000091820 UP000008792 UP000078200 UP000007266 UP000007798 UP000075880 UP000075881 UP000095301 UP000001819 UP000075882 UP000030765 UP000007062 UP000075903 UP000075883 UP000268350 UP000069272 UP000095300 UP000075884 UP000075902 UP000075920 UP000076408 UP000075886 UP000007801 UP000075900 UP000009192 UP000075901 UP000075885 UP000000673 UP000092444 UP000001070 UP000076407 UP000001292 UP000002282 UP000192223 UP000000803 UP000008711 UP000092553 UP000192221 UP000076502 UP000053825 UP000005203 UP000242457 UP000279307 UP000036403 UP000078541 UP000005205 UP000078540 UP000007755 UP000078492 UP000075809 UP000235965 UP000183832 UP000053097 UP000002358 UP000027135 UP000008237 UP000215335 UP000053105 UP000009046 UP000245037 UP000078542
UP000002320 UP000037069 UP000008820 UP000069940 UP000249989 UP000092443 UP000092460 UP000092445 UP000091820 UP000008792 UP000078200 UP000007266 UP000007798 UP000075880 UP000075881 UP000095301 UP000001819 UP000075882 UP000030765 UP000007062 UP000075903 UP000075883 UP000268350 UP000069272 UP000095300 UP000075884 UP000075902 UP000075920 UP000076408 UP000075886 UP000007801 UP000075900 UP000009192 UP000075901 UP000075885 UP000000673 UP000092444 UP000001070 UP000076407 UP000001292 UP000002282 UP000192223 UP000000803 UP000008711 UP000092553 UP000192221 UP000076502 UP000053825 UP000005203 UP000242457 UP000279307 UP000036403 UP000078541 UP000005205 UP000078540 UP000007755 UP000078492 UP000075809 UP000235965 UP000183832 UP000053097 UP000002358 UP000027135 UP000008237 UP000215335 UP000053105 UP000009046 UP000245037 UP000078542
Interpro
Gene 3D
ProteinModelPortal
H9IVG2
A0A2A4K3X3
A0A0U2LCP7
A0A3S2NTU0
A0A2H1V6Z2
A0A194PDA0
+ More
A0A1U9X1Y1 A0A0L7LDS9 A0A194RLI5 B0WNN8 A0A0L0BPS0 Q16WF1 W8C4D2 A0A182GAQ9 A0A182GJ52 A0A0K8UZL1 A0A068F7V0 A0A034VJG4 A0A0A1XRB1 A0A1A9XQZ3 A0A1B0C2N7 A0A1A9ZLW6 A0A1A9W4Y7 B4LG54 A0A1A9UP19 D2A2B8 B4N3X7 A0A182J1I1 A0A182K8Z1 A0A1I8M356 Q29DT7 A0A336L831 A0A182LJ43 A0A084WK64 A0A336LFM2 Q7Q6V8 A0A182VJS9 A0A182MVK1 A0A3B0JZQ0 A0A182FC87 A0A1I8PJ28 A0A182NGW2 A0A336M919 A0A182TE28 A0A182WII3 A0A182Y1A1 A0A182QGF7 A0A0P8XV10 A0A182RF94 B4KXZ6 A0A182SDN2 A0A182PHM2 W5JIH3 A0A1B0G7J2 B4IXF7 B3M4E3 A0A182XMX1 B4HK63 B4PKA2 A0A1W4WEL0 A0A0J9RWN7 M9MS15 P23654 B3NDH9 A0A0M4EFV3 A0A1W4UGW0 A0A154PKJ7 A0A0L7R8Y8 A0A310SCB4 A0A087ZXD6 A0A2A3EDS0 A0A3L8E242 A0A0J7KAZ7 A0A195FVE2 A0A158NW65 A0A195B871 F4WNG2 A0A195ED01 A0A151WVD4 A0A1B6MHX5 A0A0T6AYL4 A0A0C9RU64 A0A2J7R8H1 A0A1J1IXZ2 A0A026WLA0 K7IZ24 A0A067RH81 E2BI28 A0A232F7E5 A0A0N0U7U2 E0VCD0 A0A2P8Y5J5 A0A151IM92
A0A1U9X1Y1 A0A0L7LDS9 A0A194RLI5 B0WNN8 A0A0L0BPS0 Q16WF1 W8C4D2 A0A182GAQ9 A0A182GJ52 A0A0K8UZL1 A0A068F7V0 A0A034VJG4 A0A0A1XRB1 A0A1A9XQZ3 A0A1B0C2N7 A0A1A9ZLW6 A0A1A9W4Y7 B4LG54 A0A1A9UP19 D2A2B8 B4N3X7 A0A182J1I1 A0A182K8Z1 A0A1I8M356 Q29DT7 A0A336L831 A0A182LJ43 A0A084WK64 A0A336LFM2 Q7Q6V8 A0A182VJS9 A0A182MVK1 A0A3B0JZQ0 A0A182FC87 A0A1I8PJ28 A0A182NGW2 A0A336M919 A0A182TE28 A0A182WII3 A0A182Y1A1 A0A182QGF7 A0A0P8XV10 A0A182RF94 B4KXZ6 A0A182SDN2 A0A182PHM2 W5JIH3 A0A1B0G7J2 B4IXF7 B3M4E3 A0A182XMX1 B4HK63 B4PKA2 A0A1W4WEL0 A0A0J9RWN7 M9MS15 P23654 B3NDH9 A0A0M4EFV3 A0A1W4UGW0 A0A154PKJ7 A0A0L7R8Y8 A0A310SCB4 A0A087ZXD6 A0A2A3EDS0 A0A3L8E242 A0A0J7KAZ7 A0A195FVE2 A0A158NW65 A0A195B871 F4WNG2 A0A195ED01 A0A151WVD4 A0A1B6MHX5 A0A0T6AYL4 A0A0C9RU64 A0A2J7R8H1 A0A1J1IXZ2 A0A026WLA0 K7IZ24 A0A067RH81 E2BI28 A0A232F7E5 A0A0N0U7U2 E0VCD0 A0A2P8Y5J5 A0A151IM92
PDB
1C7I
E-value=3.86416e-20,
Score=244
Ontologies
GO
Topology
Subcellular location
Membrane
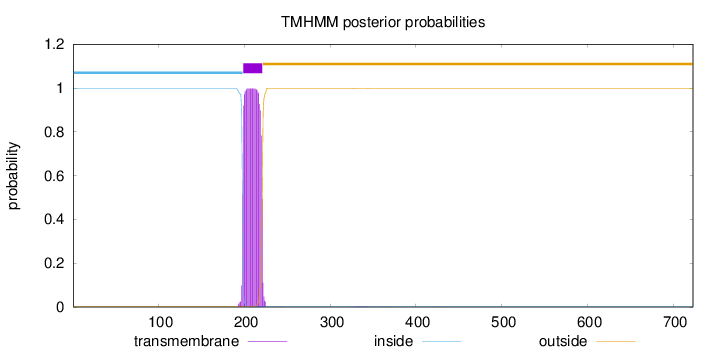
Length:
723
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.82201
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99634
inside
1 - 198
TMhelix
199 - 221
outside
222 - 723
Population Genetic Test Statistics
Pi
204.869806
Theta
176.42035
Tajima's D
0.419864
CLR
0.684586
CSRT
0.489475526223689
Interpretation
Uncertain
