Pre Gene Modal
BGIBMGA000898
Annotation
PREDICTED:_splicing_factor_3A_subunit_2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.716
Sequence
CDS
ATGGATTTTCAAAATCGCCCTGGCGGTAAAACTGGTGGAGGAGGTGTAGCGTCATGGTCGGAAAGTAACAGAGACCGGCGGGAGAGATTACGACAGCTTGCACTAGAAACAATAGATTTAAACAAAGATCCATATTTTATGAAAAATCATTTAGGGTCCTATGAATGTAAACTTTGTTTAACCCTGCATAACAATGAGGGAAGTTATTTGGCTCACACGCAAGGGAAGAAGCATCAAGCCAATTTGGCACGTAGAGCTGCAAAGGAAGCCAAAGAAGCGCCTCAACAGCTTGCCCCGGAAAAACCTAGAATTGAACCTAAGAAATTTGTGAAGATTGGTAGACCTGGTTACAGAGTAACAAAACAGAAAGATCCTGAAACTGGACAGCAGAGCTTGCTCTTCCAAGTAGATTATCCAGAGATTGCAGAGGGTGTTATACCTAGACATAGATTTATGTCTGCTTATGAACAGAAGATTGAGCCTCCAGACCGCAGATGGCAGTATTTGTTATTTGCAGCAGAGCCTTATGAAACTATTGCATTTAAAGTCCCTAGCAGAGAAGTTGAAAAGCATGATTCAAAATTCTGGACTCATTGGAACAAGGACACAAAGCAGTTCTTTTTGCAGTTTGCATTTAAGATGGATCAGTTACGCATGCCTCCCCCCCATCCCAAAATGTGGGAGGGTCATAGCATGAGGTTGCCTCCACCTCCTGGAGCACCGATGTTGGGAGTACCACCGCCACCACCATTATTACCAGTGCCCCCACCACCACCCTCTATGTAA
Protein
MDFQNRPGGKTGGGGVASWSESNRDRRERLRQLALETIDLNKDPYFMKNHLGSYECKLCLTLHNNEGSYLAHTQGKKHQANLARRAAKEAKEAPQQLAPEKPRIEPKKFVKIGRPGYRVTKQKDPETGQQSLLFQVDYPEIAEGVIPRHRFMSAYEQKIEPPDRRWQYLLFAAEPYETIAFKVPSREVEKHDSKFWTHWNKDTKQFFLQFAFKMDQLRMPPPHPKMWEGHSMRLPPPPGAPMLGVPPPPPLLPVPPPPPSM
Summary
Uniprot
A0A1E1WFD6
S4PHJ6
A0A0L7KP97
A0A194PJF2
A0A194RHC6
A0A3S2PDP5
+ More
A0A2A4JGA2 A0A0A9YEM3 A0A212FI02 A0A2R7VT21 A0A0P4VHH1 T1I7P3 A0A224XLJ9 A0A0V0GAA7 A0A069DQT2 A0A0L0CUC3 A0A336LM35 A0A1A9ZS70 A0A1L8EGY9 A0A1A9XDC7 A0A1B0BPB7 D3TMM7 A0A1I8NVI2 A0A336L8D8 A0A1A9X112 A0A1A9UQI0 A0A034VYB9 T1PGS2 A0A1B6GS13 A0A0A1WKM5 W8BQM1 A0A0K8VU76 A0A1J1IA56 T1JJG7 A0A1W4UVP6 A0A2R5LBW2 A0A3B0JP54 E2C0J2 B3M9C3 B4L0U2 A0A293LIK1 A0A0J7KFL1 Q2LYX0 B4H011 E2AVG6 B4ML36 A0A195FJ60 F4W6B0 A0A151WHQ9 A0A151IER8 A0A0K8TKY6 Q9VU15 B4PGV3 B4HG67 B3NCV2 B4QRS2 Q8I173 A0A195DMG3 A0A195AW18 A0A158P225 A0A1W4XH04 E9IV49 A0A0L7RDH6 A0A2A3EK41 A0A088AIB1 A0A310S8R3 E2C829 A0A0T6AV46 A0A026VXM1 A0A3S3P8N3 J3JY54 A0A183IXN7 B4IZM4 A0A2P8YEZ9 A0A232EG75 B7P952 V5HCS0 A0A067R2A1 K7J2Y0 A0A1Z5L8E7 A0A023FXZ9 G3MP46 A0A023GGT0 A0A2J7Q684 A0A1E1X4Z2 A0A023FK95 A0A1Q3F664 A0A077Z0V8 D2A323 A0A1Y1LZ81 A0A3P9JJ96 Q4RV48 H3DCY8 B0W7X5 A0A3S2M7U5 A0A224Z754 A0A131Z4B9 A0A131XLK1 A0A182QMG3 A0A182VNH0
A0A2A4JGA2 A0A0A9YEM3 A0A212FI02 A0A2R7VT21 A0A0P4VHH1 T1I7P3 A0A224XLJ9 A0A0V0GAA7 A0A069DQT2 A0A0L0CUC3 A0A336LM35 A0A1A9ZS70 A0A1L8EGY9 A0A1A9XDC7 A0A1B0BPB7 D3TMM7 A0A1I8NVI2 A0A336L8D8 A0A1A9X112 A0A1A9UQI0 A0A034VYB9 T1PGS2 A0A1B6GS13 A0A0A1WKM5 W8BQM1 A0A0K8VU76 A0A1J1IA56 T1JJG7 A0A1W4UVP6 A0A2R5LBW2 A0A3B0JP54 E2C0J2 B3M9C3 B4L0U2 A0A293LIK1 A0A0J7KFL1 Q2LYX0 B4H011 E2AVG6 B4ML36 A0A195FJ60 F4W6B0 A0A151WHQ9 A0A151IER8 A0A0K8TKY6 Q9VU15 B4PGV3 B4HG67 B3NCV2 B4QRS2 Q8I173 A0A195DMG3 A0A195AW18 A0A158P225 A0A1W4XH04 E9IV49 A0A0L7RDH6 A0A2A3EK41 A0A088AIB1 A0A310S8R3 E2C829 A0A0T6AV46 A0A026VXM1 A0A3S3P8N3 J3JY54 A0A183IXN7 B4IZM4 A0A2P8YEZ9 A0A232EG75 B7P952 V5HCS0 A0A067R2A1 K7J2Y0 A0A1Z5L8E7 A0A023FXZ9 G3MP46 A0A023GGT0 A0A2J7Q684 A0A1E1X4Z2 A0A023FK95 A0A1Q3F664 A0A077Z0V8 D2A323 A0A1Y1LZ81 A0A3P9JJ96 Q4RV48 H3DCY8 B0W7X5 A0A3S2M7U5 A0A224Z754 A0A131Z4B9 A0A131XLK1 A0A182QMG3 A0A182VNH0
Pubmed
23622113
26227816
26354079
25401762
26823975
22118469
+ More
27129103 26334808 26108605 20353571 25348373 25315136 25830018 24495485 20798317 17994087 15632085 21719571 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 18057021 21347285 21282665 24508170 30249741 22516182 23537049 29403074 28648823 25765539 24845553 20075255 28528879 22216098 28503490 18362917 19820115 28004739 17554307 15496914 28797301 26830274 28049606
27129103 26334808 26108605 20353571 25348373 25315136 25830018 24495485 20798317 17994087 15632085 21719571 26369729 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 18057021 21347285 21282665 24508170 30249741 22516182 23537049 29403074 28648823 25765539 24845553 20075255 28528879 22216098 28503490 18362917 19820115 28004739 17554307 15496914 28797301 26830274 28049606
EMBL
GDQN01005417
JAT85637.1
GAIX01003252
JAA89308.1
JTDY01007538
KOB65128.1
+ More
KQ459606 KPI91215.1 KQ460205 KPJ16844.1 RSAL01000084 RVE48411.1 NWSH01001481 PCG71115.1 GBHO01013010 GBRD01004396 GDHC01020245 GDHC01006703 JAG30594.1 JAG61425.1 JAP98383.1 JAQ11926.1 AGBW02008449 OWR53361.1 KK854069 PTY10652.1 GDKW01002678 JAI53917.1 ACPB03000177 GFTR01004462 JAW11964.1 GECL01001959 JAP04165.1 GBGD01002486 JAC86403.1 JRES01000005 KNC34964.1 UFQS01000052 UFQT01000052 SSW98660.1 SSX19046.1 GFDG01000861 JAV17938.1 JXJN01017879 CCAG010002320 EZ422679 ADD18955.1 UFQS01001941 UFQT01001941 SSX12745.1 SSX32187.1 GAKP01012087 JAC46865.1 KA647982 AFP62611.1 GECZ01004570 JAS65199.1 GBXI01015071 JAC99220.1 GAMC01003015 GAMC01003014 JAC03541.1 GDHF01009877 JAI42437.1 CVRI01000046 CRK97099.1 JH431165 GGLE01002888 MBY07014.1 OUUW01000002 SPP77290.1 GL451799 EFN78538.1 CH902618 EDV41136.1 CH933809 EDW19192.2 GFWV01008588 MAA33317.1 LBMM01008210 KMQ89054.1 CH379069 EAL29739.1 CH479199 EDW29588.1 GL443111 EFN62576.1 CH963847 EDW73094.1 KQ981523 KYN40406.1 GL887707 EGI70245.1 KQ983112 KYQ47348.1 KQ977858 KYM99305.1 GDAI01002802 JAI14801.1 AE014296 AY118970 AAF49876.1 AAM50830.1 CM000159 EDW94342.1 CH480815 EDW41313.1 CH954178 EDV51608.1 CM000363 CM002912 EDX10286.1 KMY99305.1 AY190958 CH940647 AAO01098.1 EDW70804.1 KQ980724 KYN14073.1 KQ976731 KYM76252.1 ADTU01006971 GL766116 EFZ15572.1 KQ414614 KOC68903.1 KZ288222 PBC32068.1 KQ768821 OAD53058.1 GL453532 EFN75902.1 LJIG01022742 KRT78952.1 KK107670 QOIP01000010 EZA48196.1 RLU17537.1 NCKU01003388 NCKU01000545 NCKU01000544 RWS07618.1 RWS15096.1 RWS15108.1 BT128178 KB632375 AEE63139.1 ERL93994.1 UZAM01011542 VDP16844.1 CH916366 EDV97799.1 PYGN01000650 PSN42832.1 NNAY01004862 OXU17328.1 ABJB010332570 ABJB010334271 ABJB010907402 DS661587 EEC03124.1 GANP01011606 GEFM01003807 JAB72862.1 JAP71989.1 KK852761 KDR17024.1 GFJQ02003308 JAW03662.1 GBBL01001740 JAC25580.1 JO843647 AEO35264.1 GBBM01002222 JAC33196.1 NEVH01017535 PNF24094.1 GFAC01004890 JAT94298.1 GBBK01003419 JAC21063.1 GFDL01012026 JAV23019.1 HG805850 CDW53363.1 KQ971338 EFA01964.1 GEZM01046888 JAV76976.1 CAAE01014992 CAG07734.1 DS231856 EDS38437.1 CM012453 RVE61806.1 GFPF01012025 MAA23171.1 GEDV01002308 JAP86249.1 GEFH01000622 JAP67959.1 AXCN02001143
KQ459606 KPI91215.1 KQ460205 KPJ16844.1 RSAL01000084 RVE48411.1 NWSH01001481 PCG71115.1 GBHO01013010 GBRD01004396 GDHC01020245 GDHC01006703 JAG30594.1 JAG61425.1 JAP98383.1 JAQ11926.1 AGBW02008449 OWR53361.1 KK854069 PTY10652.1 GDKW01002678 JAI53917.1 ACPB03000177 GFTR01004462 JAW11964.1 GECL01001959 JAP04165.1 GBGD01002486 JAC86403.1 JRES01000005 KNC34964.1 UFQS01000052 UFQT01000052 SSW98660.1 SSX19046.1 GFDG01000861 JAV17938.1 JXJN01017879 CCAG010002320 EZ422679 ADD18955.1 UFQS01001941 UFQT01001941 SSX12745.1 SSX32187.1 GAKP01012087 JAC46865.1 KA647982 AFP62611.1 GECZ01004570 JAS65199.1 GBXI01015071 JAC99220.1 GAMC01003015 GAMC01003014 JAC03541.1 GDHF01009877 JAI42437.1 CVRI01000046 CRK97099.1 JH431165 GGLE01002888 MBY07014.1 OUUW01000002 SPP77290.1 GL451799 EFN78538.1 CH902618 EDV41136.1 CH933809 EDW19192.2 GFWV01008588 MAA33317.1 LBMM01008210 KMQ89054.1 CH379069 EAL29739.1 CH479199 EDW29588.1 GL443111 EFN62576.1 CH963847 EDW73094.1 KQ981523 KYN40406.1 GL887707 EGI70245.1 KQ983112 KYQ47348.1 KQ977858 KYM99305.1 GDAI01002802 JAI14801.1 AE014296 AY118970 AAF49876.1 AAM50830.1 CM000159 EDW94342.1 CH480815 EDW41313.1 CH954178 EDV51608.1 CM000363 CM002912 EDX10286.1 KMY99305.1 AY190958 CH940647 AAO01098.1 EDW70804.1 KQ980724 KYN14073.1 KQ976731 KYM76252.1 ADTU01006971 GL766116 EFZ15572.1 KQ414614 KOC68903.1 KZ288222 PBC32068.1 KQ768821 OAD53058.1 GL453532 EFN75902.1 LJIG01022742 KRT78952.1 KK107670 QOIP01000010 EZA48196.1 RLU17537.1 NCKU01003388 NCKU01000545 NCKU01000544 RWS07618.1 RWS15096.1 RWS15108.1 BT128178 KB632375 AEE63139.1 ERL93994.1 UZAM01011542 VDP16844.1 CH916366 EDV97799.1 PYGN01000650 PSN42832.1 NNAY01004862 OXU17328.1 ABJB010332570 ABJB010334271 ABJB010907402 DS661587 EEC03124.1 GANP01011606 GEFM01003807 JAB72862.1 JAP71989.1 KK852761 KDR17024.1 GFJQ02003308 JAW03662.1 GBBL01001740 JAC25580.1 JO843647 AEO35264.1 GBBM01002222 JAC33196.1 NEVH01017535 PNF24094.1 GFAC01004890 JAT94298.1 GBBK01003419 JAC21063.1 GFDL01012026 JAV23019.1 HG805850 CDW53363.1 KQ971338 EFA01964.1 GEZM01046888 JAV76976.1 CAAE01014992 CAG07734.1 DS231856 EDS38437.1 CM012453 RVE61806.1 GFPF01012025 MAA23171.1 GEDV01002308 JAP86249.1 GEFH01000622 JAP67959.1 AXCN02001143
Proteomes
UP000037510
UP000053268
UP000053240
UP000283053
UP000218220
UP000007151
+ More
UP000015103 UP000037069 UP000092445 UP000092443 UP000092460 UP000092444 UP000095300 UP000091820 UP000078200 UP000095301 UP000183832 UP000192221 UP000268350 UP000008237 UP000007801 UP000009192 UP000036403 UP000001819 UP000008744 UP000000311 UP000007798 UP000078541 UP000007755 UP000075809 UP000078542 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000008792 UP000078492 UP000078540 UP000005205 UP000192223 UP000053825 UP000242457 UP000005203 UP000053097 UP000279307 UP000285301 UP000030742 UP000050793 UP000001070 UP000245037 UP000215335 UP000001555 UP000027135 UP000002358 UP000235965 UP000030665 UP000007266 UP000265200 UP000007303 UP000002320 UP000075886 UP000075903
UP000015103 UP000037069 UP000092445 UP000092443 UP000092460 UP000092444 UP000095300 UP000091820 UP000078200 UP000095301 UP000183832 UP000192221 UP000268350 UP000008237 UP000007801 UP000009192 UP000036403 UP000001819 UP000008744 UP000000311 UP000007798 UP000078541 UP000007755 UP000075809 UP000078542 UP000000803 UP000002282 UP000001292 UP000008711 UP000000304 UP000008792 UP000078492 UP000078540 UP000005205 UP000192223 UP000053825 UP000242457 UP000005203 UP000053097 UP000279307 UP000285301 UP000030742 UP000050793 UP000001070 UP000245037 UP000215335 UP000001555 UP000027135 UP000002358 UP000235965 UP000030665 UP000007266 UP000265200 UP000007303 UP000002320 UP000075886 UP000075903
PRIDE
Interpro
ProteinModelPortal
A0A1E1WFD6
S4PHJ6
A0A0L7KP97
A0A194PJF2
A0A194RHC6
A0A3S2PDP5
+ More
A0A2A4JGA2 A0A0A9YEM3 A0A212FI02 A0A2R7VT21 A0A0P4VHH1 T1I7P3 A0A224XLJ9 A0A0V0GAA7 A0A069DQT2 A0A0L0CUC3 A0A336LM35 A0A1A9ZS70 A0A1L8EGY9 A0A1A9XDC7 A0A1B0BPB7 D3TMM7 A0A1I8NVI2 A0A336L8D8 A0A1A9X112 A0A1A9UQI0 A0A034VYB9 T1PGS2 A0A1B6GS13 A0A0A1WKM5 W8BQM1 A0A0K8VU76 A0A1J1IA56 T1JJG7 A0A1W4UVP6 A0A2R5LBW2 A0A3B0JP54 E2C0J2 B3M9C3 B4L0U2 A0A293LIK1 A0A0J7KFL1 Q2LYX0 B4H011 E2AVG6 B4ML36 A0A195FJ60 F4W6B0 A0A151WHQ9 A0A151IER8 A0A0K8TKY6 Q9VU15 B4PGV3 B4HG67 B3NCV2 B4QRS2 Q8I173 A0A195DMG3 A0A195AW18 A0A158P225 A0A1W4XH04 E9IV49 A0A0L7RDH6 A0A2A3EK41 A0A088AIB1 A0A310S8R3 E2C829 A0A0T6AV46 A0A026VXM1 A0A3S3P8N3 J3JY54 A0A183IXN7 B4IZM4 A0A2P8YEZ9 A0A232EG75 B7P952 V5HCS0 A0A067R2A1 K7J2Y0 A0A1Z5L8E7 A0A023FXZ9 G3MP46 A0A023GGT0 A0A2J7Q684 A0A1E1X4Z2 A0A023FK95 A0A1Q3F664 A0A077Z0V8 D2A323 A0A1Y1LZ81 A0A3P9JJ96 Q4RV48 H3DCY8 B0W7X5 A0A3S2M7U5 A0A224Z754 A0A131Z4B9 A0A131XLK1 A0A182QMG3 A0A182VNH0
A0A2A4JGA2 A0A0A9YEM3 A0A212FI02 A0A2R7VT21 A0A0P4VHH1 T1I7P3 A0A224XLJ9 A0A0V0GAA7 A0A069DQT2 A0A0L0CUC3 A0A336LM35 A0A1A9ZS70 A0A1L8EGY9 A0A1A9XDC7 A0A1B0BPB7 D3TMM7 A0A1I8NVI2 A0A336L8D8 A0A1A9X112 A0A1A9UQI0 A0A034VYB9 T1PGS2 A0A1B6GS13 A0A0A1WKM5 W8BQM1 A0A0K8VU76 A0A1J1IA56 T1JJG7 A0A1W4UVP6 A0A2R5LBW2 A0A3B0JP54 E2C0J2 B3M9C3 B4L0U2 A0A293LIK1 A0A0J7KFL1 Q2LYX0 B4H011 E2AVG6 B4ML36 A0A195FJ60 F4W6B0 A0A151WHQ9 A0A151IER8 A0A0K8TKY6 Q9VU15 B4PGV3 B4HG67 B3NCV2 B4QRS2 Q8I173 A0A195DMG3 A0A195AW18 A0A158P225 A0A1W4XH04 E9IV49 A0A0L7RDH6 A0A2A3EK41 A0A088AIB1 A0A310S8R3 E2C829 A0A0T6AV46 A0A026VXM1 A0A3S3P8N3 J3JY54 A0A183IXN7 B4IZM4 A0A2P8YEZ9 A0A232EG75 B7P952 V5HCS0 A0A067R2A1 K7J2Y0 A0A1Z5L8E7 A0A023FXZ9 G3MP46 A0A023GGT0 A0A2J7Q684 A0A1E1X4Z2 A0A023FK95 A0A1Q3F664 A0A077Z0V8 D2A323 A0A1Y1LZ81 A0A3P9JJ96 Q4RV48 H3DCY8 B0W7X5 A0A3S2M7U5 A0A224Z754 A0A131Z4B9 A0A131XLK1 A0A182QMG3 A0A182VNH0
PDB
6QX9
E-value=3.7877e-85,
Score=800
Ontologies
GO
Topology
Subcellular location
Nucleus
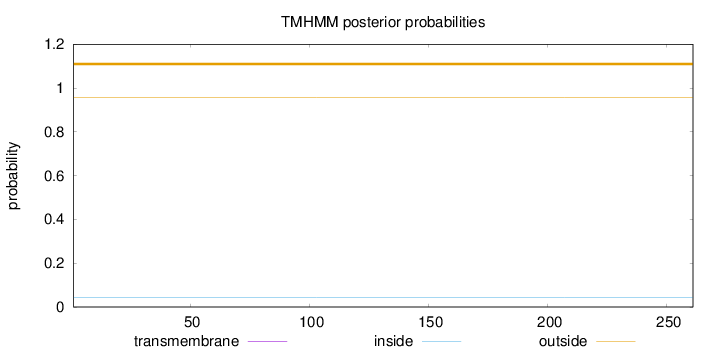
Length:
261
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04202
outside
1 - 261
Population Genetic Test Statistics
Pi
125.191638
Theta
183.787053
Tajima's D
-1.011289
CLR
336.175083
CSRT
0.131443427828609
Interpretation
Uncertain
