Gene
KWMTBOMO07772 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001243
Annotation
ribosomal_protein_S10_[Bombyx_mori]
Full name
40S ribosomal protein S10
+ More
40S ribosomal protein S10b
40S ribosomal protein S10b
Location in the cell
Cytoplasmic Reliability : 1.942 Mitochondrial Reliability : 1.92
Sequence
CDS
ATGTTGATGCCCAAACAAAACCGTGTTGCTATTTATGAGTACCTCTTTAAAGAGGGAGTCATGGTGGCCAAAAAAGACTATCATGCACCGAAGCATACTGAACTAGAGAAGATTCCTAACCTCCAAGTTATCAAAGCTATGCAATCACTCAAGTCCAGAGGTTATGTCAAAGAACAGTTTGCCTGGAGGCACTTTTACTGGTACCTAACCAATGAGGGTATTGAATACTTGAGAATATTCTTGCACCTTCCTCCTGAAATTGTGCCTGCAACACTTAAGCGCTCAGTACGTACAGAGACAGTACGCCGTGGCCCTGTTGGTCGTCCTGATGCCCCAGCCCGCTCTGCTGAGGACAGATCTGCATACAGACGTACTCCTGCTGCCCCTGGTGTTGCACCTCATGATAAAAAGGCTGATGTTGGTCCAGGCTCAGCTGATCTTGAATTTAAGGGAGGCTATGGACGTGGCAGGCCTGCTTCATAA
Protein
MLMPKQNRVAIYEYLFKEGVMVAKKDYHAPKHTELEKIPNLQVIKAMQSLKSRGYVKEQFAWRHFYWYLTNEGIEYLRIFLHLPPEIVPATLKRSVRTETVRRGPVGRPDAPARSAEDRSAYRRTPAAPGVAPHDKKADVGPGSADLEFKGGYGRGRPAS
Summary
Similarity
Belongs to the eukaryotic ribosomal protein eS10 family.
Keywords
Cytoplasm
Ribonucleoprotein
Ribosomal protein
Complete proteome
Reference proteome
Feature
chain 40S ribosomal protein S10
Uniprot
Q5UAN4
H9IVG3
D1LYQ8
A0A3G1T1M0
S4PBB2
A0A194RG98
+ More
I4DIM1 A0A2H1V6U7 Q962R9 I4DMC5 E7DZ52 A0A2A4JHR2 A0A212FI05 Q5MGM0 A7KCY7 G0ZEH1 A0A1E1W3Y5 A0A194PEU2 A0A0L7KNM3 A2IAD6 Q6EUZ6 V5G8N7 D1M831 Q4GXT1 D2A360 Q56FF9 A0A1B1ZGH0 A0A1Y1K1J2 A0A0L7RIL9 A0A0M9A5Y0 A0A151IQ96 A0A195FES7 A0A151XD37 Q4GXS9 A0A232EVN5 A0A195DWY1 A0A195B7E1 A0A158NGX1 Q6EUZ4 K7IZL3 F4WZU5 Q6EUZ5 V9IFV9 A0A088AHT2 A0A0C9QMC3 A0A2A3EA23 A2I3Y9 A0A154PS66 A0A240SYN0 A0A1B0GLM3 A0A1L8E3A7 U5EUI2 E9ITT7 A0A1L8E394 A0A0J7K5Z5 A0A026W9V6 E2BVL8 A0A1B0CJS2 E2B1C9 B7ZWP7 D4G7H1 C0H6Z8 M9NEQ9 Q9VWG3 T1PFK3 B4R7R9 B4I775 A0A1I8P6M1 A0A1L8EHC6 Q29IV5 W8CBA8 A0A3B0KAS4 A0A034VLR1 B4PZG3 A0A0A1XPI9 B4NDM2 A0A0R1EHM5 B3NVQ0 A0A0C9RB89 A0A1B6IRS6 A0A0L0BT67 B3MQA2 A0A2C9NEF3 A0A1A9Y8N9 A0A1A9ZBC6 B4L809 A0A1W4VU69 B4MAS4 A0A1B6KIU3 A0A109NIA2 T1D546 B4JME5 A0A0U2UPU2 A0A084VFR4 R4V0G7 A0A087UYA3 A0A023FWR8 A0A067QVN3 A0A0C9S0V6 A0A1D1YH56
I4DIM1 A0A2H1V6U7 Q962R9 I4DMC5 E7DZ52 A0A2A4JHR2 A0A212FI05 Q5MGM0 A7KCY7 G0ZEH1 A0A1E1W3Y5 A0A194PEU2 A0A0L7KNM3 A2IAD6 Q6EUZ6 V5G8N7 D1M831 Q4GXT1 D2A360 Q56FF9 A0A1B1ZGH0 A0A1Y1K1J2 A0A0L7RIL9 A0A0M9A5Y0 A0A151IQ96 A0A195FES7 A0A151XD37 Q4GXS9 A0A232EVN5 A0A195DWY1 A0A195B7E1 A0A158NGX1 Q6EUZ4 K7IZL3 F4WZU5 Q6EUZ5 V9IFV9 A0A088AHT2 A0A0C9QMC3 A0A2A3EA23 A2I3Y9 A0A154PS66 A0A240SYN0 A0A1B0GLM3 A0A1L8E3A7 U5EUI2 E9ITT7 A0A1L8E394 A0A0J7K5Z5 A0A026W9V6 E2BVL8 A0A1B0CJS2 E2B1C9 B7ZWP7 D4G7H1 C0H6Z8 M9NEQ9 Q9VWG3 T1PFK3 B4R7R9 B4I775 A0A1I8P6M1 A0A1L8EHC6 Q29IV5 W8CBA8 A0A3B0KAS4 A0A034VLR1 B4PZG3 A0A0A1XPI9 B4NDM2 A0A0R1EHM5 B3NVQ0 A0A0C9RB89 A0A1B6IRS6 A0A0L0BT67 B3MQA2 A0A2C9NEF3 A0A1A9Y8N9 A0A1A9ZBC6 B4L809 A0A1W4VU69 B4MAS4 A0A1B6KIU3 A0A109NIA2 T1D546 B4JME5 A0A0U2UPU2 A0A084VFR4 R4V0G7 A0A087UYA3 A0A023FWR8 A0A067QVN3 A0A0C9S0V6 A0A1D1YH56
Pubmed
19121390
23622113
26354079
22651552
14668217
22118469
+ More
16023793 17603110 26227816 17437641 18362917 19820115 27464714 28004739 28648823 21347285 20075255 21719571 21282665 24508170 30249741 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17994087 15632085 24495485 25348373 17550304 25830018 18057021 26108605 24330624 26621068 24438588 24845553 26131772
16023793 17603110 26227816 17437641 18362917 19820115 27464714 28004739 28648823 21347285 20075255 21719571 21282665 24508170 30249741 20798317 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17994087 15632085 24495485 25348373 17550304 25830018 18057021 26108605 24330624 26621068 24438588 24845553 26131772
EMBL
AY769324
AAV34866.1
BABH01001043
GU084332
ACY95365.1
MG992442
+ More
AXY94880.1 GAIX01005997 JAA86563.1 KQ460205 KPJ16843.1 AK401139 BAM17761.1 ODYU01000982 SOQ36558.1 AF400207 AK402443 BAM19065.1 HQ424755 ADT80699.1 NWSH01001481 PCG71124.1 AGBW02008449 OWR53360.1 AY829766 AAV91380.1 EF207982 ABS57455.1 JF265049 AEL28880.1 GDQN01009392 JAT81662.1 KQ459606 KPI91214.1 JTDY01007873 KOB64892.1 EF179450 ABM55456.1 AJ783875 CAH04323.1 GALX01001996 JAB66470.1 GU120455 ACY71305.1 AM048942 CAJ17179.1 KQ971338 EFA02264.1 AY961541 AAX62443.1 KU756281 ANX99824.1 GEZM01098160 JAV53970.1 KQ414583 KOC70710.1 KQ435727 KOX77900.1 KQ976780 KYN08328.1 KQ981625 KYN39180.1 KQ982294 KYQ58275.1 AM048944 CAJ17181.1 NNAY01001980 OXU22385.1 KQ980204 KYN17227.1 KQ976574 KYM80167.1 ADTU01015300 AJ783877 CAH04325.1 GL888480 EGI60370.1 AJ783876 CAH04324.1 JR045461 AEY59983.1 GBYB01004669 JAG74436.1 KZ288310 PBC28577.1 EF070492 ABM55558.1 KQ435119 KZC14749.1 AJVK01027443 AJWK01033653 GFDF01000877 JAV13207.1 GANO01001458 JAB58413.1 GL765695 EFZ16016.1 GFDF01000874 JAV13210.1 LBMM01013371 KMQ85669.1 KK107314 QOIP01000012 EZA52830.1 RLU15866.1 GL450875 EFN80288.1 AJWK01014905 GL444841 EFN60529.1 BT050589 ACJ49254.1 BT122123 ADE06729.1 BT058104 ACN09817.1 AE014298 AFH07470.1 AHN59915.1 AY070896 KA647454 AFP62083.1 CM000366 EDX18402.1 CH480823 EDW56173.1 GFDG01000708 JAV18091.1 CH379063 EAL32548.2 GAMC01000629 JAC05927.1 OUUW01000034 SPP89822.1 GAKP01014706 JAC44246.1 CM000162 EDX02119.1 GBXI01001356 JAD12936.1 CH964239 EDW81844.1 KRK06570.1 CH954180 EDV46715.1 KQS30097.1 GBYB01013749 JAG83516.1 GECU01018088 JAS89618.1 JRES01001383 KNC23242.1 CH902621 EDV44528.1 CCAG010013939 CH933814 EDW05584.1 CH940655 EDW66333.1 GEBQ01028632 JAT11345.1 KC507307 AHB12430.1 GALA01000688 JAA94164.1 CH916371 EDV92470.1 KT755156 ALS04990.1 ATLV01012458 KE524793 KFB36808.1 KC632273 AGM32087.1 KK122254 KFM82342.1 GBBL01002180 JAC25140.1 KK853320 KDR08546.1 GBZX01002386 JAG90354.1 GDJX01013967 JAT53969.1
AXY94880.1 GAIX01005997 JAA86563.1 KQ460205 KPJ16843.1 AK401139 BAM17761.1 ODYU01000982 SOQ36558.1 AF400207 AK402443 BAM19065.1 HQ424755 ADT80699.1 NWSH01001481 PCG71124.1 AGBW02008449 OWR53360.1 AY829766 AAV91380.1 EF207982 ABS57455.1 JF265049 AEL28880.1 GDQN01009392 JAT81662.1 KQ459606 KPI91214.1 JTDY01007873 KOB64892.1 EF179450 ABM55456.1 AJ783875 CAH04323.1 GALX01001996 JAB66470.1 GU120455 ACY71305.1 AM048942 CAJ17179.1 KQ971338 EFA02264.1 AY961541 AAX62443.1 KU756281 ANX99824.1 GEZM01098160 JAV53970.1 KQ414583 KOC70710.1 KQ435727 KOX77900.1 KQ976780 KYN08328.1 KQ981625 KYN39180.1 KQ982294 KYQ58275.1 AM048944 CAJ17181.1 NNAY01001980 OXU22385.1 KQ980204 KYN17227.1 KQ976574 KYM80167.1 ADTU01015300 AJ783877 CAH04325.1 GL888480 EGI60370.1 AJ783876 CAH04324.1 JR045461 AEY59983.1 GBYB01004669 JAG74436.1 KZ288310 PBC28577.1 EF070492 ABM55558.1 KQ435119 KZC14749.1 AJVK01027443 AJWK01033653 GFDF01000877 JAV13207.1 GANO01001458 JAB58413.1 GL765695 EFZ16016.1 GFDF01000874 JAV13210.1 LBMM01013371 KMQ85669.1 KK107314 QOIP01000012 EZA52830.1 RLU15866.1 GL450875 EFN80288.1 AJWK01014905 GL444841 EFN60529.1 BT050589 ACJ49254.1 BT122123 ADE06729.1 BT058104 ACN09817.1 AE014298 AFH07470.1 AHN59915.1 AY070896 KA647454 AFP62083.1 CM000366 EDX18402.1 CH480823 EDW56173.1 GFDG01000708 JAV18091.1 CH379063 EAL32548.2 GAMC01000629 JAC05927.1 OUUW01000034 SPP89822.1 GAKP01014706 JAC44246.1 CM000162 EDX02119.1 GBXI01001356 JAD12936.1 CH964239 EDW81844.1 KRK06570.1 CH954180 EDV46715.1 KQS30097.1 GBYB01013749 JAG83516.1 GECU01018088 JAS89618.1 JRES01001383 KNC23242.1 CH902621 EDV44528.1 CCAG010013939 CH933814 EDW05584.1 CH940655 EDW66333.1 GEBQ01028632 JAT11345.1 KC507307 AHB12430.1 GALA01000688 JAA94164.1 CH916371 EDV92470.1 KT755156 ALS04990.1 ATLV01012458 KE524793 KFB36808.1 KC632273 AGM32087.1 KK122254 KFM82342.1 GBBL01002180 JAC25140.1 KK853320 KDR08546.1 GBZX01002386 JAG90354.1 GDJX01013967 JAT53969.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000053268
UP000037510
+ More
UP000007266 UP000053825 UP000053105 UP000078542 UP000078541 UP000075809 UP000215335 UP000078492 UP000078540 UP000005205 UP000002358 UP000007755 UP000005203 UP000242457 UP000076502 UP000092462 UP000092461 UP000036403 UP000053097 UP000279307 UP000008237 UP000000311 UP000000803 UP000095301 UP000000304 UP000001292 UP000095300 UP000001819 UP000268350 UP000002282 UP000007798 UP000008711 UP000037069 UP000007801 UP000092444 UP000092443 UP000092445 UP000009192 UP000192221 UP000008792 UP000001070 UP000030765 UP000054359 UP000027135
UP000007266 UP000053825 UP000053105 UP000078542 UP000078541 UP000075809 UP000215335 UP000078492 UP000078540 UP000005205 UP000002358 UP000007755 UP000005203 UP000242457 UP000076502 UP000092462 UP000092461 UP000036403 UP000053097 UP000279307 UP000008237 UP000000311 UP000000803 UP000095301 UP000000304 UP000001292 UP000095300 UP000001819 UP000268350 UP000002282 UP000007798 UP000008711 UP000037069 UP000007801 UP000092444 UP000092443 UP000092445 UP000009192 UP000192221 UP000008792 UP000001070 UP000030765 UP000054359 UP000027135
Pfam
PF03501 S10_plectin
Gene 3D
ProteinModelPortal
Q5UAN4
H9IVG3
D1LYQ8
A0A3G1T1M0
S4PBB2
A0A194RG98
+ More
I4DIM1 A0A2H1V6U7 Q962R9 I4DMC5 E7DZ52 A0A2A4JHR2 A0A212FI05 Q5MGM0 A7KCY7 G0ZEH1 A0A1E1W3Y5 A0A194PEU2 A0A0L7KNM3 A2IAD6 Q6EUZ6 V5G8N7 D1M831 Q4GXT1 D2A360 Q56FF9 A0A1B1ZGH0 A0A1Y1K1J2 A0A0L7RIL9 A0A0M9A5Y0 A0A151IQ96 A0A195FES7 A0A151XD37 Q4GXS9 A0A232EVN5 A0A195DWY1 A0A195B7E1 A0A158NGX1 Q6EUZ4 K7IZL3 F4WZU5 Q6EUZ5 V9IFV9 A0A088AHT2 A0A0C9QMC3 A0A2A3EA23 A2I3Y9 A0A154PS66 A0A240SYN0 A0A1B0GLM3 A0A1L8E3A7 U5EUI2 E9ITT7 A0A1L8E394 A0A0J7K5Z5 A0A026W9V6 E2BVL8 A0A1B0CJS2 E2B1C9 B7ZWP7 D4G7H1 C0H6Z8 M9NEQ9 Q9VWG3 T1PFK3 B4R7R9 B4I775 A0A1I8P6M1 A0A1L8EHC6 Q29IV5 W8CBA8 A0A3B0KAS4 A0A034VLR1 B4PZG3 A0A0A1XPI9 B4NDM2 A0A0R1EHM5 B3NVQ0 A0A0C9RB89 A0A1B6IRS6 A0A0L0BT67 B3MQA2 A0A2C9NEF3 A0A1A9Y8N9 A0A1A9ZBC6 B4L809 A0A1W4VU69 B4MAS4 A0A1B6KIU3 A0A109NIA2 T1D546 B4JME5 A0A0U2UPU2 A0A084VFR4 R4V0G7 A0A087UYA3 A0A023FWR8 A0A067QVN3 A0A0C9S0V6 A0A1D1YH56
I4DIM1 A0A2H1V6U7 Q962R9 I4DMC5 E7DZ52 A0A2A4JHR2 A0A212FI05 Q5MGM0 A7KCY7 G0ZEH1 A0A1E1W3Y5 A0A194PEU2 A0A0L7KNM3 A2IAD6 Q6EUZ6 V5G8N7 D1M831 Q4GXT1 D2A360 Q56FF9 A0A1B1ZGH0 A0A1Y1K1J2 A0A0L7RIL9 A0A0M9A5Y0 A0A151IQ96 A0A195FES7 A0A151XD37 Q4GXS9 A0A232EVN5 A0A195DWY1 A0A195B7E1 A0A158NGX1 Q6EUZ4 K7IZL3 F4WZU5 Q6EUZ5 V9IFV9 A0A088AHT2 A0A0C9QMC3 A0A2A3EA23 A2I3Y9 A0A154PS66 A0A240SYN0 A0A1B0GLM3 A0A1L8E3A7 U5EUI2 E9ITT7 A0A1L8E394 A0A0J7K5Z5 A0A026W9V6 E2BVL8 A0A1B0CJS2 E2B1C9 B7ZWP7 D4G7H1 C0H6Z8 M9NEQ9 Q9VWG3 T1PFK3 B4R7R9 B4I775 A0A1I8P6M1 A0A1L8EHC6 Q29IV5 W8CBA8 A0A3B0KAS4 A0A034VLR1 B4PZG3 A0A0A1XPI9 B4NDM2 A0A0R1EHM5 B3NVQ0 A0A0C9RB89 A0A1B6IRS6 A0A0L0BT67 B3MQA2 A0A2C9NEF3 A0A1A9Y8N9 A0A1A9ZBC6 B4L809 A0A1W4VU69 B4MAS4 A0A1B6KIU3 A0A109NIA2 T1D546 B4JME5 A0A0U2UPU2 A0A084VFR4 R4V0G7 A0A087UYA3 A0A023FWR8 A0A067QVN3 A0A0C9S0V6 A0A1D1YH56
PDB
4V6W
E-value=1.83933e-47,
Score=472
Ontologies
PANTHER
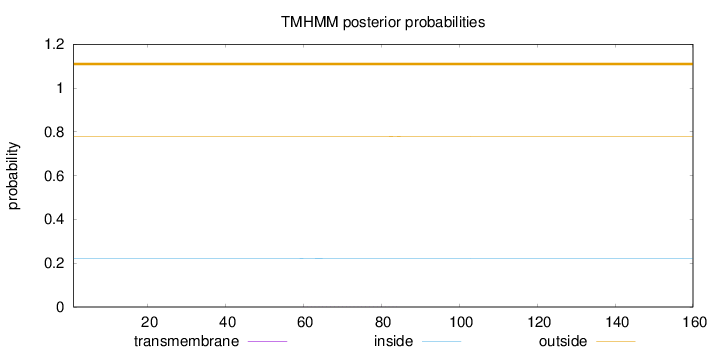
Topology
Subcellular location
Cytoplasm
Length:
160
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01657
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.22135
outside
1 - 160
Population Genetic Test Statistics
Pi
237.370416
Theta
210.266516
Tajima's D
0.312028
CLR
0
CSRT
0.456227188640568
Interpretation
Uncertain
