Gene
KWMTBOMO07771 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000897
Annotation
PREDICTED:_NADH_dehydrogenase_[ubiquinone]_1_alpha_subcomplex_subunit_9?_mitochondrial_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.161 Mitochondrial Reliability : 1.778
Sequence
CDS
ATGGCTGCGATAGCACTAAAAACTCAAGCTACTAGCAAATTACTTCATTTGAATGGGTCTATGAGTGTTGTGTACATCAAAGCCGCTAACTACAGTTCGGATCGTAAACCCAACCTTGCAGCATATAAGCGAGGTACTGGTGGCCGCAGTAGCTTTAACGGCATCGTAGCCACAGTTTTCGGTTGCACCGGATTTGTCGGACGCTATGTGTGCAACAAATTGGGAAAAATTGGTACCCAGTTAATTTTACCATACAGAGGCGATTTCTATGATGCCCAAAGGTTGAAAGTGTGCGGAGATTTAGGCCAGGTCCTGTTTACTCCTTACCATCTCCTTGATGAGGAGTCTATTGCAAAAGCGGTCCGATACTCTAATGTTGTCATCAATCTAGTTGGACGAGATTATGAAACTAAGAATTTCAAATACAATGATGTTCACGTTGATGGTGTAAGAAGAATTGCCAGAATCTGCAGAGAAGAAGGAGTTGAGAGATTCATTCATCTGTCTTATTTAAATGCTGAAGAACATCCGAAACCTCTTGTCCTGAAGAAACCGTCTGCTTGGAAAATAAGCAAGTATTTGGGAGAGTGTGCAGTCAGAGAGGAATACCCAACTGCCACTATTATCCGAGCATCAGACATCTATGGTTCCGAAGACAGATTTTTGAGGTCATTAGTTAATAAGATGCGATCTCACAGCAACCTGATGCCCCTCTATAAAAACGGATTAGCGACTGTGAAACAGCCTGTGTTTGTGTCGGACGTCGCTCAGGGTATCGTGAACGCCGCGCGCGACGACGACACTAAATGTGAAGTCTACCAAGCCGTTGGGCCAAAACGCTACTTACTTGCCGATTTGGTTGATTGGTTTTACAAACTAATGAGGAAGGACGAAAAATGGGGTGGATACATTCGCTACGACATGAAGTACGACCCGATACTGCCCCTGAAAGTTGCTTTGGTTAATGCCATTTCACCAGCTTATCCACTCGGAAATCTTCACTGGGAAGGAATTGAAAGGGAAGCTACTTCAGATAATGTGGTGATCGGCGTGCCCACCCTTGAGGATTTGGGCGTCACCTTGACGCACATGGAAGACCAGGTTCCGTGGGAACTGAAACCTTTTCGCGCCCATCAGTACTACATGGATCGGCTTGGAGAGTTCCCGAAACCAGACCCTCCACCAGTTTACTCAGCTTGA
Protein
MAAIALKTQATSKLLHLNGSMSVVYIKAANYSSDRKPNLAAYKRGTGGRSSFNGIVATVFGCTGFVGRYVCNKLGKIGTQLILPYRGDFYDAQRLKVCGDLGQVLFTPYHLLDEESIAKAVRYSNVVINLVGRDYETKNFKYNDVHVDGVRRIARICREEGVERFIHLSYLNAEEHPKPLVLKKPSAWKISKYLGECAVREEYPTATIIRASDIYGSEDRFLRSLVNKMRSHSNLMPLYKNGLATVKQPVFVSDVAQGIVNAARDDDTKCEVYQAVGPKRYLLADLVDWFYKLMRKDEKWGGYIRYDMKYDPILPLKVALVNAISPAYPLGNLHWEGIEREATSDNVVIGVPTLEDLGVTLTHMEDQVPWELKPFRAHQYYMDRLGEFPKPDPPPVYSA
Summary
Uniprot
H9IUG7
A0A194RGX8
I4DK06
S4PL76
A0A2A4JI70
A0A212FI25
+ More
A0A2H1V6Y3 A0A0L0BMX8 A0A2J7QIX5 I4DRB5 A0A1I8P848 A0A1L8EE22 U5EV20 A0A0K8TTJ5 A0A182IKN8 A0A1Q3FI43 A0A067RSM5 A0A1I8MLV6 B0X750 A0A336L840 A0A2M4BQH8 T1PFJ6 A0A182JQU4 A0A2M3Z2L7 A0A2M4BQD4 A0A1L8E0I7 A0A2P8ZAT2 T1E7B9 A0A182X0H8 A0A182UXZ9 A0A182KV97 A0A182TUB9 A0A182I5R7 A0A2M4A972 A0A182YIX4 A0A1L8E0F2 W5JDN0 A0A182W9J6 Q7Q229 A0A1L8E0R4 A0A182REI7 W8B7U2 A0A182P1X3 A0A182FVV8 A0A0A1WQM3 A0A182QNV9 A0A182M0S8 B4LGC0 B4KY50 A0A084VB40 A0A023ERK2 Q16VZ4 A0A023EQE5 A0A034VW24 A0A182NV78 A0A1A9UM39 A0A1Y1KG50 A0A1A9WH60 D3TQE0 A0A1B0G2S6 A0A1A9ZDK7 B4N6L5 A0A1A9YLS9 A0A1B0C3X0 B3M9B1 B4IYY9 B4IUH0 A0A1B0AFU1 B3NIN3 Q29DC3 A0A024E3A5 B4PET2 B4IAC1 A0A1W4W6S1 A0A0J9RYS1 D2A3E2 B4H7N4 Q9VPE2 A0A0M4EC20 A0A3B0JVT2 A0A1B0G1G5 A0A1J1HX26 V5GWM3 A0A0P6DC76 A0A0P5G5L8 A0A1B6DM59 A0A0P5KYJ3 A0A0P5TSG6 A0A0P5F4A9 A0A0P5NYG7 A0A0P5N7K0 A0A0P5Y9R2 A0A0P5ARE0 A0A0P5J9Q0 A0A0P5AKQ8 A0A0P5HPJ5 A0A0P6FL69 A0A0P5WRD9 A0A1B0DP10 A0A0P6FYH9
A0A2H1V6Y3 A0A0L0BMX8 A0A2J7QIX5 I4DRB5 A0A1I8P848 A0A1L8EE22 U5EV20 A0A0K8TTJ5 A0A182IKN8 A0A1Q3FI43 A0A067RSM5 A0A1I8MLV6 B0X750 A0A336L840 A0A2M4BQH8 T1PFJ6 A0A182JQU4 A0A2M3Z2L7 A0A2M4BQD4 A0A1L8E0I7 A0A2P8ZAT2 T1E7B9 A0A182X0H8 A0A182UXZ9 A0A182KV97 A0A182TUB9 A0A182I5R7 A0A2M4A972 A0A182YIX4 A0A1L8E0F2 W5JDN0 A0A182W9J6 Q7Q229 A0A1L8E0R4 A0A182REI7 W8B7U2 A0A182P1X3 A0A182FVV8 A0A0A1WQM3 A0A182QNV9 A0A182M0S8 B4LGC0 B4KY50 A0A084VB40 A0A023ERK2 Q16VZ4 A0A023EQE5 A0A034VW24 A0A182NV78 A0A1A9UM39 A0A1Y1KG50 A0A1A9WH60 D3TQE0 A0A1B0G2S6 A0A1A9ZDK7 B4N6L5 A0A1A9YLS9 A0A1B0C3X0 B3M9B1 B4IYY9 B4IUH0 A0A1B0AFU1 B3NIN3 Q29DC3 A0A024E3A5 B4PET2 B4IAC1 A0A1W4W6S1 A0A0J9RYS1 D2A3E2 B4H7N4 Q9VPE2 A0A0M4EC20 A0A3B0JVT2 A0A1B0G1G5 A0A1J1HX26 V5GWM3 A0A0P6DC76 A0A0P5G5L8 A0A1B6DM59 A0A0P5KYJ3 A0A0P5TSG6 A0A0P5F4A9 A0A0P5NYG7 A0A0P5N7K0 A0A0P5Y9R2 A0A0P5ARE0 A0A0P5J9Q0 A0A0P5AKQ8 A0A0P5HPJ5 A0A0P6FL69 A0A0P5WRD9 A0A1B0DP10 A0A0P6FYH9
Pubmed
19121390
26354079
22651552
23622113
22118469
26108605
+ More
26369729 24845553 25315136 29403074 20966253 25244985 20920257 23761445 12364791 24495485 25830018 17994087 24438588 24945155 26483478 17510324 25348373 28004739 20353571 17550304 15632085 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
26369729 24845553 25315136 29403074 20966253 25244985 20920257 23761445 12364791 24495485 25830018 17994087 24438588 24945155 26483478 17510324 25348373 28004739 20353571 17550304 15632085 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01001043
KQ460205
KPJ16842.1
AK401624
KQ459606
BAM18246.1
+ More
KPI91213.1 GAIX01004195 JAA88365.1 NWSH01001481 PCG71123.1 AGBW02008449 OWR53359.1 ODYU01000982 SOQ36559.1 JRES01001711 KNC20629.1 NEVH01013570 PNF28532.1 AK405052 BAM20455.1 GFDG01001905 JAV16894.1 GANO01002068 JAB57803.1 GDAI01000127 JAI17476.1 GFDL01007809 JAV27236.1 KK852444 KDR23820.1 DS232438 EDS41744.1 UFQS01000218 UFQS01001264 UFQT01000218 UFQT01001264 SSX01495.1 SSX10035.1 SSX21875.1 GGFJ01006040 MBW55181.1 KA647449 AFP62078.1 GGFM01001937 MBW22688.1 GGFJ01006042 MBW55183.1 GFDF01001831 JAV12253.1 PYGN01000121 PSN53607.1 GAMD01003182 JAA98408.1 APCN01003489 GGFK01003847 MBW37168.1 GFDF01001876 JAV12208.1 ADMH02001521 ETN62206.1 AAAB01008979 EAA13714.3 GFDF01001830 JAV12254.1 GAMC01013447 JAB93108.1 GBXI01013120 JAD01172.1 AXCN02001805 AXCM01001817 CH940647 EDW70449.1 CH933809 EDW18752.1 ATLV01005273 KE524276 KFB35184.1 JXUM01157484 JXUM01157485 JXUM01157486 GAPW01001790 KQ572762 JAC11808.1 KXJ68000.1 CH477579 EAT38719.1 JXUM01070465 JXUM01070466 GAPW01001791 KQ562608 JAC11807.1 KXJ75488.1 GAKP01012897 JAC46055.1 GEZM01087931 JAV58386.1 EZ423642 ADD19918.1 CCAG010015567 CH964161 EDW80004.1 JXJN01025135 CH902618 EDV40095.1 CH916366 EDV97697.1 CH891906 EDX00034.1 CH954178 EDV52529.1 CH379070 EAL30491.1 BT150449 AHZ64223.1 CM000159 EDW95156.1 CH480826 EDW44234.1 CM002912 KMZ00812.1 KQ971338 EFA02874.1 CH479219 EDW34674.1 AE014296 BT150453 AAF51613.1 AGB94809.1 AIG12978.1 CP012525 ALC45116.1 OUUW01000002 SPP76841.1 CCAG010012985 CVRI01000033 CRK92632.1 GALX01003813 JAB64653.1 GDIQ01082806 JAN11931.1 GDIP01254837 GDIP01200297 GDIQ01267820 GDIP01100499 GDIP01045524 GDIQ01053580 LRGB01001579 JAI68564.1 JAJ83904.1 KZS11557.1 GEDC01010566 JAS26732.1 GDIQ01185621 JAK66104.1 GDIP01122205 JAL81509.1 GDIQ01265926 JAJ85798.1 GDIQ01157374 JAK94351.1 GDIQ01157375 JAK94350.1 GDIP01060858 JAM42857.1 GDIP01195046 JAJ28356.1 GDIQ01267083 GDIQ01203265 JAK48460.1 GDIP01211342 GDIP01147051 JAJ12060.1 GDIQ01247705 JAK04020.1 GDIQ01045877 JAN48860.1 GDIP01082698 JAM21017.1 AJVK01017960 GDIQ01043771 JAN50966.1
KPI91213.1 GAIX01004195 JAA88365.1 NWSH01001481 PCG71123.1 AGBW02008449 OWR53359.1 ODYU01000982 SOQ36559.1 JRES01001711 KNC20629.1 NEVH01013570 PNF28532.1 AK405052 BAM20455.1 GFDG01001905 JAV16894.1 GANO01002068 JAB57803.1 GDAI01000127 JAI17476.1 GFDL01007809 JAV27236.1 KK852444 KDR23820.1 DS232438 EDS41744.1 UFQS01000218 UFQS01001264 UFQT01000218 UFQT01001264 SSX01495.1 SSX10035.1 SSX21875.1 GGFJ01006040 MBW55181.1 KA647449 AFP62078.1 GGFM01001937 MBW22688.1 GGFJ01006042 MBW55183.1 GFDF01001831 JAV12253.1 PYGN01000121 PSN53607.1 GAMD01003182 JAA98408.1 APCN01003489 GGFK01003847 MBW37168.1 GFDF01001876 JAV12208.1 ADMH02001521 ETN62206.1 AAAB01008979 EAA13714.3 GFDF01001830 JAV12254.1 GAMC01013447 JAB93108.1 GBXI01013120 JAD01172.1 AXCN02001805 AXCM01001817 CH940647 EDW70449.1 CH933809 EDW18752.1 ATLV01005273 KE524276 KFB35184.1 JXUM01157484 JXUM01157485 JXUM01157486 GAPW01001790 KQ572762 JAC11808.1 KXJ68000.1 CH477579 EAT38719.1 JXUM01070465 JXUM01070466 GAPW01001791 KQ562608 JAC11807.1 KXJ75488.1 GAKP01012897 JAC46055.1 GEZM01087931 JAV58386.1 EZ423642 ADD19918.1 CCAG010015567 CH964161 EDW80004.1 JXJN01025135 CH902618 EDV40095.1 CH916366 EDV97697.1 CH891906 EDX00034.1 CH954178 EDV52529.1 CH379070 EAL30491.1 BT150449 AHZ64223.1 CM000159 EDW95156.1 CH480826 EDW44234.1 CM002912 KMZ00812.1 KQ971338 EFA02874.1 CH479219 EDW34674.1 AE014296 BT150453 AAF51613.1 AGB94809.1 AIG12978.1 CP012525 ALC45116.1 OUUW01000002 SPP76841.1 CCAG010012985 CVRI01000033 CRK92632.1 GALX01003813 JAB64653.1 GDIQ01082806 JAN11931.1 GDIP01254837 GDIP01200297 GDIQ01267820 GDIP01100499 GDIP01045524 GDIQ01053580 LRGB01001579 JAI68564.1 JAJ83904.1 KZS11557.1 GEDC01010566 JAS26732.1 GDIQ01185621 JAK66104.1 GDIP01122205 JAL81509.1 GDIQ01265926 JAJ85798.1 GDIQ01157374 JAK94351.1 GDIQ01157375 JAK94350.1 GDIP01060858 JAM42857.1 GDIP01195046 JAJ28356.1 GDIQ01267083 GDIQ01203265 JAK48460.1 GDIP01211342 GDIP01147051 JAJ12060.1 GDIQ01247705 JAK04020.1 GDIQ01045877 JAN48860.1 GDIP01082698 JAM21017.1 AJVK01017960 GDIQ01043771 JAN50966.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037069
+ More
UP000235965 UP000095300 UP000075880 UP000027135 UP000095301 UP000002320 UP000075881 UP000245037 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000076408 UP000000673 UP000075920 UP000007062 UP000075900 UP000075885 UP000069272 UP000075886 UP000075883 UP000008792 UP000009192 UP000030765 UP000069940 UP000249989 UP000008820 UP000075884 UP000078200 UP000091820 UP000092444 UP000092445 UP000007798 UP000092443 UP000092460 UP000007801 UP000001070 UP000002282 UP000008711 UP000001819 UP000001292 UP000192221 UP000007266 UP000008744 UP000000803 UP000092553 UP000268350 UP000183832 UP000076858 UP000092462
UP000235965 UP000095300 UP000075880 UP000027135 UP000095301 UP000002320 UP000075881 UP000245037 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000076408 UP000000673 UP000075920 UP000007062 UP000075900 UP000075885 UP000069272 UP000075886 UP000075883 UP000008792 UP000009192 UP000030765 UP000069940 UP000249989 UP000008820 UP000075884 UP000078200 UP000091820 UP000092444 UP000092445 UP000007798 UP000092443 UP000092460 UP000007801 UP000001070 UP000002282 UP000008711 UP000001819 UP000001292 UP000192221 UP000007266 UP000008744 UP000000803 UP000092553 UP000268350 UP000183832 UP000076858 UP000092462
Pfam
PF01370 Epimerase
Interpro
Gene 3D
ProteinModelPortal
H9IUG7
A0A194RGX8
I4DK06
S4PL76
A0A2A4JI70
A0A212FI25
+ More
A0A2H1V6Y3 A0A0L0BMX8 A0A2J7QIX5 I4DRB5 A0A1I8P848 A0A1L8EE22 U5EV20 A0A0K8TTJ5 A0A182IKN8 A0A1Q3FI43 A0A067RSM5 A0A1I8MLV6 B0X750 A0A336L840 A0A2M4BQH8 T1PFJ6 A0A182JQU4 A0A2M3Z2L7 A0A2M4BQD4 A0A1L8E0I7 A0A2P8ZAT2 T1E7B9 A0A182X0H8 A0A182UXZ9 A0A182KV97 A0A182TUB9 A0A182I5R7 A0A2M4A972 A0A182YIX4 A0A1L8E0F2 W5JDN0 A0A182W9J6 Q7Q229 A0A1L8E0R4 A0A182REI7 W8B7U2 A0A182P1X3 A0A182FVV8 A0A0A1WQM3 A0A182QNV9 A0A182M0S8 B4LGC0 B4KY50 A0A084VB40 A0A023ERK2 Q16VZ4 A0A023EQE5 A0A034VW24 A0A182NV78 A0A1A9UM39 A0A1Y1KG50 A0A1A9WH60 D3TQE0 A0A1B0G2S6 A0A1A9ZDK7 B4N6L5 A0A1A9YLS9 A0A1B0C3X0 B3M9B1 B4IYY9 B4IUH0 A0A1B0AFU1 B3NIN3 Q29DC3 A0A024E3A5 B4PET2 B4IAC1 A0A1W4W6S1 A0A0J9RYS1 D2A3E2 B4H7N4 Q9VPE2 A0A0M4EC20 A0A3B0JVT2 A0A1B0G1G5 A0A1J1HX26 V5GWM3 A0A0P6DC76 A0A0P5G5L8 A0A1B6DM59 A0A0P5KYJ3 A0A0P5TSG6 A0A0P5F4A9 A0A0P5NYG7 A0A0P5N7K0 A0A0P5Y9R2 A0A0P5ARE0 A0A0P5J9Q0 A0A0P5AKQ8 A0A0P5HPJ5 A0A0P6FL69 A0A0P5WRD9 A0A1B0DP10 A0A0P6FYH9
A0A2H1V6Y3 A0A0L0BMX8 A0A2J7QIX5 I4DRB5 A0A1I8P848 A0A1L8EE22 U5EV20 A0A0K8TTJ5 A0A182IKN8 A0A1Q3FI43 A0A067RSM5 A0A1I8MLV6 B0X750 A0A336L840 A0A2M4BQH8 T1PFJ6 A0A182JQU4 A0A2M3Z2L7 A0A2M4BQD4 A0A1L8E0I7 A0A2P8ZAT2 T1E7B9 A0A182X0H8 A0A182UXZ9 A0A182KV97 A0A182TUB9 A0A182I5R7 A0A2M4A972 A0A182YIX4 A0A1L8E0F2 W5JDN0 A0A182W9J6 Q7Q229 A0A1L8E0R4 A0A182REI7 W8B7U2 A0A182P1X3 A0A182FVV8 A0A0A1WQM3 A0A182QNV9 A0A182M0S8 B4LGC0 B4KY50 A0A084VB40 A0A023ERK2 Q16VZ4 A0A023EQE5 A0A034VW24 A0A182NV78 A0A1A9UM39 A0A1Y1KG50 A0A1A9WH60 D3TQE0 A0A1B0G2S6 A0A1A9ZDK7 B4N6L5 A0A1A9YLS9 A0A1B0C3X0 B3M9B1 B4IYY9 B4IUH0 A0A1B0AFU1 B3NIN3 Q29DC3 A0A024E3A5 B4PET2 B4IAC1 A0A1W4W6S1 A0A0J9RYS1 D2A3E2 B4H7N4 Q9VPE2 A0A0M4EC20 A0A3B0JVT2 A0A1B0G1G5 A0A1J1HX26 V5GWM3 A0A0P6DC76 A0A0P5G5L8 A0A1B6DM59 A0A0P5KYJ3 A0A0P5TSG6 A0A0P5F4A9 A0A0P5NYG7 A0A0P5N7K0 A0A0P5Y9R2 A0A0P5ARE0 A0A0P5J9Q0 A0A0P5AKQ8 A0A0P5HPJ5 A0A0P6FL69 A0A0P5WRD9 A0A1B0DP10 A0A0P6FYH9
PDB
5LNK
E-value=2.73355e-68,
Score=657
Ontologies
PATHWAY
GO
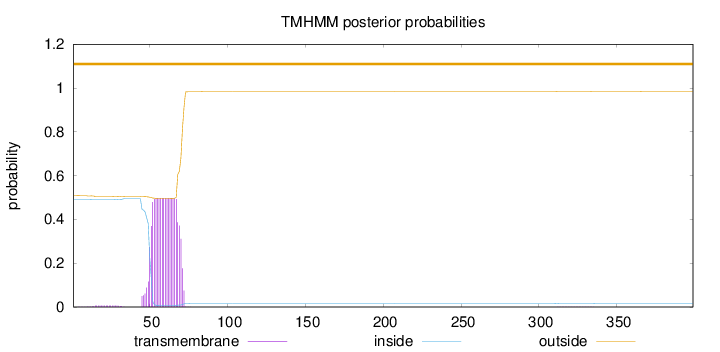
Topology
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.39847
Exp number, first 60 AAs:
5.58534
Total prob of N-in:
0.49106
outside
1 - 399
Population Genetic Test Statistics
Pi
200.417525
Theta
169.44916
Tajima's D
0.446918
CLR
0.006042
CSRT
0.500474976251187
Interpretation
Uncertain
