Gene
KWMTBOMO07769
Pre Gene Modal
BGIBMGA001245
Annotation
PREDICTED:_spondin-1-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.98
Sequence
CDS
ATGCACGAGACGGAAGCGTTGAGGTTTTTATTGGTCTTGGTAGTTTCTTTTGAGAATGCGATATTTGTTGCTGGTGAGTCCTTGGCTAAGTGTGAACAGGCTCCGGCGCACGCGCAGCCCACCGATGATCATTTCAGCTTGCGTATATCGGGCGAACCAGATTTGTTTATACCAGGAGAGCTGTATACCGTTACGCTCCGAGGGGCTAACAGCGGTCAAGGCCCCTATCCGTTTACCGGCTTCAAGATATGGGCTGAAGTCGCGAGTGATTTTTCAAAAGACGAAACAGAGCAAGGAGACCTGAGAGCAACAGCTGACCCTCACCAGCCGCTGGGAGTATTCCAGGCTTACGATGCTCAGACTAAACTTCATGAATCCTGTGCTCCAGCCGTCGATAATGCTACTGCCCACCCTAAGACAGAAGTGCAGGTAATATGGAATGCACCTTTAAGTGTGAGAGGAGTTTGCGTGCGGCTGTCTGCTCGTGTCGGTCCCACGCTGACTGCAAATAAATGGTCTGTTTTGGCTCGTAAGATCTGCCCCACTTCGGCTGGACCTTCTAGCCTGAATCGACAACCACCCATTGTTGAGCCCTGCTGTGCTTGTGATGAAGCAAAATATGAGGTAAATTTCGAAGGAATGTGGTCTCGCAACACCCATCCTCGTGAGTTTCCTCCGGAGAATGCTCATGCCCATTTTGGTGATATAATTGGCGCCTCACATACTGCACAGTACAGGGTCTGGCAAGAAGGTCGGGTAGCGACGACTGGTTTAAGGCGTCTCGCCGATGACGGCTCTACCACAGCCCTCGAAAAAGAATTAAAAGCAGAGAGTGATCATATCCGTACGATAATCAAAGCCCGTGGTATATCGTGGCAGCAAGTGACGGGAACTGGCATTCCAAGTACGTTTGCTGTGTTTCGTGTAGACGCCAAGCACCATCTGGTATCTTTGGCTGCCAAATTAGCTCCTTCGCCAGACTGGATCGTAGGAGTTTCGGCGTTGGAGCTCTGCAACGTGAATTGTACATGGAGAACGTCGGCCTCACTGTCGTTGTACCCTTACGATGCTGGTACAGATAATGGCATAAGTTATACTTCTCGCCGTATGCCAACGATTCCAGCGGCGCCCGTACGAGCCCTTAGGACGGATTGGCCTCGCGATGTGCGCTCACCGTTTTTCAGCACGACCGGGGAAATGAGACCCTTTGCGCGCGTTCGATTAACCAGATTGCGTTTATATGAGAAGAGCTGTGATGTCACAGAGCCCGGAACGGATCAGAACGTTCAAGAAGGTGTCGGTGCGGGAGGCTGGTGCGCGACACACGGGTGGGGTCCGTGGGGTCCGTGCAGCGTGACGTGCGGTGCGGGCCGCGCCTCCCGCCAGCGCGACTACGTCTGGCCGGCGCGGGCCCACGCTGAGGCTTGTCGCGTACAACTCACAGATTACAAACACTGCCACGGACCGAGGATGCATTGCCGAGCTGTATCGGAATACGAGCCGGACCCAGCGGAGTCGAGCGGCCCGTGTGCGATGTCTCCGTGGTCGGAGTGGTCCCCGTGCGACGGGTGCGGGGTGCGGGCCCGCACGCGGCACTACCTCGCCCCGCGAGCGTACAAACGATGTCACATCGGCTTCCGCGCCCGCACTATACTCAGCCAGGCCATGCCCTGCGCCGAGGGACCGTGTGAACAACCACCGCCCCGATACGCGAACATGTCGAGTTTCGATTGGTTTTTCGTGGACAATCCTAGAGGAGAGTGTAGAGTGACGTCATGGAGCGACTGGTCGCCGTGCTCGGCGCGGTGCGGGCGCGGCCGCCGACTGCGCACGCGCGTGTACGTAGTGCGGGAGACGCGCGTGCAACAGCTCATGTCGAAACGATTGCTGGCTCAGTGGAATCAACGTTTTGCCGAATTGCAGAACTTGGAAGTTGCTACAGAAAACGTAACCTCAGAGGACCCAGAAATTGAAAATCTCGTAGAGGAACATCTGGAACGATGCCAGTTCACTTTGACTCAACAAGAGGCCTTTTGCGACGGAGAAGACAGAAGCTGCATCAATTTTAATGACTCTTCAAAAATCTGCAAGCTACCGATGTCGGTCGGCCATTGTCGCAGGTATCAGGAGCGTTGGTTCTTTGACTGGCCCCGCAAGTCCTGTGAACCGTTCGGATACTCCGGCTGCGGAGGGAACGACAATAATTTCCGTGACAGAGATGAGTGTTTGAGCGTATGTCACAAGAACACCTCGGTAGGAAAGAAATCGGAATCTGTGAGCGAAACGAGAATTAAAAAAATGAAGGCGACCCCTTCATCTGATGACAACGAGGTCATGTTGAATGACAAGCCAACGGTGCGCGATTCAATGAAGTGTGCAACTGATTAG
Protein
MHETEALRFLLVLVVSFENAIFVAGESLAKCEQAPAHAQPTDDHFSLRISGEPDLFIPGELYTVTLRGANSGQGPYPFTGFKIWAEVASDFSKDETEQGDLRATADPHQPLGVFQAYDAQTKLHESCAPAVDNATAHPKTEVQVIWNAPLSVRGVCVRLSARVGPTLTANKWSVLARKICPTSAGPSSLNRQPPIVEPCCACDEAKYEVNFEGMWSRNTHPREFPPENAHAHFGDIIGASHTAQYRVWQEGRVATTGLRRLADDGSTTALEKELKAESDHIRTIIKARGISWQQVTGTGIPSTFAVFRVDAKHHLVSLAAKLAPSPDWIVGVSALELCNVNCTWRTSASLSLYPYDAGTDNGISYTSRRMPTIPAAPVRALRTDWPRDVRSPFFSTTGEMRPFARVRLTRLRLYEKSCDVTEPGTDQNVQEGVGAGGWCATHGWGPWGPCSVTCGAGRASRQRDYVWPARAHAEACRVQLTDYKHCHGPRMHCRAVSEYEPDPAESSGPCAMSPWSEWSPCDGCGVRARTRHYLAPRAYKRCHIGFRARTILSQAMPCAEGPCEQPPPRYANMSSFDWFFVDNPRGECRVTSWSDWSPCSARCGRGRRLRTRVYVVRETRVQQLMSKRLLAQWNQRFAELQNLEVATENVTSEDPEIENLVEEHLERCQFTLTQQEAFCDGEDRSCINFNDSSKICKLPMSVGHCRRYQERWFFDWPRKSCEPFGYSGCGGNDNNFRDRDECLSVCHKNTSVGKKSESVSETRIKKMKATPSSDDNEVMLNDKPTVRDSMKCATD
Summary
Uniprot
A0A2A4JGL1
A0A2H1V6W0
A0A3S2TK83
A0A194PD14
A0A194RR62
E0VCJ0
+ More
A0A2J7PR62 U5EYT8 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A1Q3FPJ2 A0A1Q3FP78 A0A182R5C7 A0A1Q3FPU6 Q170L4 A0A2M4CJ21 A0A182UWY8 A0A182TS53 A0A182XNC5 A0A182JZY3 A0A182KSL1 A0A182PBS9 A0A182MYL4 A0A182QL14 A0A336LL51 A0A182HQ46 A0A182FU59 A0A182W944 A0A182M9X5 A0A182Y1R5 A0A0K8STY7 A0A0P5PPB9 A0A182JBS3 A0A0A9Z7L3 A0A0P5G5S6 A0A1L8E3B7 A0A1L8E3B6 A0A0P5H4F8 A0A0P5CIT8 T1JBH1 Q7Q082 A0A0P5Z9I4 A0A0N8CXC5 A0A0P5SZF3 A0A0P4ZG86 A0A0P5IU67 A0A0P6CTJ0 A0A0N8CEJ0 A0A0P5QQF6 A0A0P5WQD1 A0A0N8ARA9 A0A1B0EWS7 A0A0P5TWQ4 A0A0N8AJJ8 A0A0P6I4P9 A0A0P5SCA1 A0A0P6GS18 A0A0P5MPN1 A0A0N8BMI8 A0A0P5S9E5 A0A0P5E4B7 A0A0P5UAZ6 A0A0P6DCY0 A0A0P5EMH8 A0A0N8BPV7 A0A0P5XD67 B0WNK9 A0A0P6EAL7 A0A0P5EU30 A0A0P5KVB3 A0A0P5MWG3 A0A0P5KSU2 A0A0P6FLY3 A0A023F2I8 A0A084WHT0 A0A0P5K323 A0A0P5T3I4 A0A0N8D582 A0A0P6FNM0 T1K1N8 A0A0P5UGB3 A0A067RIU1 A0A182T7Y8 A0A0P6G7S6 A0A0P5A418 A0A0P5TFB0 A0A0P5AL01 A0A0P5EHT2 W5J1Y7 A0A0P5ULQ1 A0A2M4AIB3 A0A224XEU1 A0A2M4BE31 A0A182GYU9 A0A0P5I8X5 A0A0P5R1Y0 E2BUL2
A0A2J7PR62 U5EYT8 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A1Q3FPJ2 A0A1Q3FP78 A0A182R5C7 A0A1Q3FPU6 Q170L4 A0A2M4CJ21 A0A182UWY8 A0A182TS53 A0A182XNC5 A0A182JZY3 A0A182KSL1 A0A182PBS9 A0A182MYL4 A0A182QL14 A0A336LL51 A0A182HQ46 A0A182FU59 A0A182W944 A0A182M9X5 A0A182Y1R5 A0A0K8STY7 A0A0P5PPB9 A0A182JBS3 A0A0A9Z7L3 A0A0P5G5S6 A0A1L8E3B7 A0A1L8E3B6 A0A0P5H4F8 A0A0P5CIT8 T1JBH1 Q7Q082 A0A0P5Z9I4 A0A0N8CXC5 A0A0P5SZF3 A0A0P4ZG86 A0A0P5IU67 A0A0P6CTJ0 A0A0N8CEJ0 A0A0P5QQF6 A0A0P5WQD1 A0A0N8ARA9 A0A1B0EWS7 A0A0P5TWQ4 A0A0N8AJJ8 A0A0P6I4P9 A0A0P5SCA1 A0A0P6GS18 A0A0P5MPN1 A0A0N8BMI8 A0A0P5S9E5 A0A0P5E4B7 A0A0P5UAZ6 A0A0P6DCY0 A0A0P5EMH8 A0A0N8BPV7 A0A0P5XD67 B0WNK9 A0A0P6EAL7 A0A0P5EU30 A0A0P5KVB3 A0A0P5MWG3 A0A0P5KSU2 A0A0P6FLY3 A0A023F2I8 A0A084WHT0 A0A0P5K323 A0A0P5T3I4 A0A0N8D582 A0A0P6FNM0 T1K1N8 A0A0P5UGB3 A0A067RIU1 A0A182T7Y8 A0A0P6G7S6 A0A0P5A418 A0A0P5TFB0 A0A0P5AL01 A0A0P5EHT2 W5J1Y7 A0A0P5ULQ1 A0A2M4AIB3 A0A224XEU1 A0A2M4BE31 A0A182GYU9 A0A0P5I8X5 A0A0P5R1Y0 E2BUL2
Pubmed
EMBL
NWSH01001481
PCG71121.1
ODYU01000982
SOQ36561.1
RSAL01000084
RVE48406.1
+ More
KQ459606 KPI91211.1 KQ459700 KPJ20368.1 DS235053 EEB11096.1 NEVH01022633 PNF18806.1 GANO01000077 JAB59794.1 PNF18807.1 PNF18808.1 PNF18809.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 GFDL01005537 JAV29508.1 CH477472 EAT40381.1 GGFL01000700 MBW64878.1 AXCN02001040 UFQT01000038 SSX18435.1 APCN01003292 AXCM01008352 GBRD01009167 JAG56654.1 GDIQ01125376 JAL26350.1 GBHO01005824 GDHC01010700 JAG37780.1 JAQ07929.1 GDIQ01267746 JAJ83978.1 GFDF01000867 JAV13217.1 GFDF01000866 JAV13218.1 GDIQ01233233 GDIQ01119073 GDIP01095690 JAK18492.1 JAM08025.1 GDIP01185568 JAJ37834.1 JH432010 AAAB01008986 EAA00346.3 GDIP01046852 JAM56863.1 GDIP01089647 JAM14068.1 GDIP01134289 JAL69425.1 GDIP01213072 JAJ10330.1 GDIQ01210190 JAK41535.1 GDIQ01087539 JAN07198.1 GDIP01139527 JAL64187.1 GDIQ01113789 JAL37937.1 GDIP01095689 JAM08026.1 GDIQ01250587 GDIQ01233232 GDIQ01163152 GDIQ01132337 JAK01138.1 AJWK01006547 AJWK01006548 AJWK01006549 GDIP01138432 JAL65282.1 GDIQ01269475 JAJ82249.1 GDIQ01009616 JAN85121.1 GDIP01141635 JAL62079.1 GDIQ01029118 JAN65619.1 GDIQ01156604 JAK95121.1 GDIQ01265527 GDIQ01233231 GDIQ01203000 GDIQ01188747 GDIQ01163155 GDIQ01160359 GDIQ01119072 JAK88570.1 GDIP01142732 JAL60982.1 GDIP01147227 JAJ76175.1 GDIP01132845 JAL70869.1 GDIQ01079590 GDIQ01048851 JAN15147.1 GDIQ01267301 JAJ84423.1 GDIQ01156603 JAK95122.1 GDIP01073851 JAM29864.1 DS232012 EDS31836.1 GDIQ01065551 JAN29186.1 GDIQ01267300 GDIQ01197652 JAJ84424.1 GDIQ01180097 JAK71628.1 GDIQ01158613 JAK93112.1 GDIQ01180096 JAK71629.1 GDIQ01045511 JAN49226.1 GBBI01003473 JAC15239.1 ATLV01023878 KE525347 KFB49774.1 GDIQ01215998 GDIQ01112476 JAK35727.1 GDIP01131532 JAL72182.1 GDIP01067591 JAM36124.1 GDIQ01045510 JAN49227.1 CAEY01001353 GDIP01113440 JAL90274.1 KK852606 KDR20370.1 GDIQ01037345 JAN57392.1 GDIP01208772 JAJ14630.1 GDIP01127234 JAL76480.1 GDIP01197830 JAJ25572.1 GDIQ01270118 JAJ81606.1 ADMH02002215 ETN57751.1 GDIP01128621 JAL75093.1 GGFK01007202 MBW40523.1 GFTR01008128 JAW08298.1 GGFJ01002112 MBW51253.1 JXUM01098242 KQ564405 KXJ72335.1 GDIQ01217146 JAK34579.1 GDIQ01108980 JAL42746.1 GL450721 EFN80621.1
KQ459606 KPI91211.1 KQ459700 KPJ20368.1 DS235053 EEB11096.1 NEVH01022633 PNF18806.1 GANO01000077 JAB59794.1 PNF18807.1 PNF18808.1 PNF18809.1 GFDL01005525 JAV29520.1 GFDL01005644 JAV29401.1 GFDL01005537 JAV29508.1 CH477472 EAT40381.1 GGFL01000700 MBW64878.1 AXCN02001040 UFQT01000038 SSX18435.1 APCN01003292 AXCM01008352 GBRD01009167 JAG56654.1 GDIQ01125376 JAL26350.1 GBHO01005824 GDHC01010700 JAG37780.1 JAQ07929.1 GDIQ01267746 JAJ83978.1 GFDF01000867 JAV13217.1 GFDF01000866 JAV13218.1 GDIQ01233233 GDIQ01119073 GDIP01095690 JAK18492.1 JAM08025.1 GDIP01185568 JAJ37834.1 JH432010 AAAB01008986 EAA00346.3 GDIP01046852 JAM56863.1 GDIP01089647 JAM14068.1 GDIP01134289 JAL69425.1 GDIP01213072 JAJ10330.1 GDIQ01210190 JAK41535.1 GDIQ01087539 JAN07198.1 GDIP01139527 JAL64187.1 GDIQ01113789 JAL37937.1 GDIP01095689 JAM08026.1 GDIQ01250587 GDIQ01233232 GDIQ01163152 GDIQ01132337 JAK01138.1 AJWK01006547 AJWK01006548 AJWK01006549 GDIP01138432 JAL65282.1 GDIQ01269475 JAJ82249.1 GDIQ01009616 JAN85121.1 GDIP01141635 JAL62079.1 GDIQ01029118 JAN65619.1 GDIQ01156604 JAK95121.1 GDIQ01265527 GDIQ01233231 GDIQ01203000 GDIQ01188747 GDIQ01163155 GDIQ01160359 GDIQ01119072 JAK88570.1 GDIP01142732 JAL60982.1 GDIP01147227 JAJ76175.1 GDIP01132845 JAL70869.1 GDIQ01079590 GDIQ01048851 JAN15147.1 GDIQ01267301 JAJ84423.1 GDIQ01156603 JAK95122.1 GDIP01073851 JAM29864.1 DS232012 EDS31836.1 GDIQ01065551 JAN29186.1 GDIQ01267300 GDIQ01197652 JAJ84424.1 GDIQ01180097 JAK71628.1 GDIQ01158613 JAK93112.1 GDIQ01180096 JAK71629.1 GDIQ01045511 JAN49226.1 GBBI01003473 JAC15239.1 ATLV01023878 KE525347 KFB49774.1 GDIQ01215998 GDIQ01112476 JAK35727.1 GDIP01131532 JAL72182.1 GDIP01067591 JAM36124.1 GDIQ01045510 JAN49227.1 CAEY01001353 GDIP01113440 JAL90274.1 KK852606 KDR20370.1 GDIQ01037345 JAN57392.1 GDIP01208772 JAJ14630.1 GDIP01127234 JAL76480.1 GDIP01197830 JAJ25572.1 GDIQ01270118 JAJ81606.1 ADMH02002215 ETN57751.1 GDIP01128621 JAL75093.1 GGFK01007202 MBW40523.1 GFTR01008128 JAW08298.1 GGFJ01002112 MBW51253.1 JXUM01098242 KQ564405 KXJ72335.1 GDIQ01217146 JAK34579.1 GDIQ01108980 JAL42746.1 GL450721 EFN80621.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000009046
UP000235965
+ More
UP000075900 UP000008820 UP000075903 UP000075902 UP000076407 UP000075881 UP000075882 UP000075885 UP000075884 UP000075886 UP000075840 UP000069272 UP000075920 UP000075883 UP000076408 UP000075880 UP000007062 UP000092461 UP000002320 UP000030765 UP000015104 UP000027135 UP000075901 UP000000673 UP000069940 UP000249989 UP000008237
UP000075900 UP000008820 UP000075903 UP000075902 UP000076407 UP000075881 UP000075882 UP000075885 UP000075884 UP000075886 UP000075840 UP000069272 UP000075920 UP000075883 UP000076408 UP000075880 UP000007062 UP000092461 UP000002320 UP000030765 UP000015104 UP000027135 UP000075901 UP000000673 UP000069940 UP000249989 UP000008237
Interpro
ProteinModelPortal
A0A2A4JGL1
A0A2H1V6W0
A0A3S2TK83
A0A194PD14
A0A194RR62
E0VCJ0
+ More
A0A2J7PR62 U5EYT8 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A1Q3FPJ2 A0A1Q3FP78 A0A182R5C7 A0A1Q3FPU6 Q170L4 A0A2M4CJ21 A0A182UWY8 A0A182TS53 A0A182XNC5 A0A182JZY3 A0A182KSL1 A0A182PBS9 A0A182MYL4 A0A182QL14 A0A336LL51 A0A182HQ46 A0A182FU59 A0A182W944 A0A182M9X5 A0A182Y1R5 A0A0K8STY7 A0A0P5PPB9 A0A182JBS3 A0A0A9Z7L3 A0A0P5G5S6 A0A1L8E3B7 A0A1L8E3B6 A0A0P5H4F8 A0A0P5CIT8 T1JBH1 Q7Q082 A0A0P5Z9I4 A0A0N8CXC5 A0A0P5SZF3 A0A0P4ZG86 A0A0P5IU67 A0A0P6CTJ0 A0A0N8CEJ0 A0A0P5QQF6 A0A0P5WQD1 A0A0N8ARA9 A0A1B0EWS7 A0A0P5TWQ4 A0A0N8AJJ8 A0A0P6I4P9 A0A0P5SCA1 A0A0P6GS18 A0A0P5MPN1 A0A0N8BMI8 A0A0P5S9E5 A0A0P5E4B7 A0A0P5UAZ6 A0A0P6DCY0 A0A0P5EMH8 A0A0N8BPV7 A0A0P5XD67 B0WNK9 A0A0P6EAL7 A0A0P5EU30 A0A0P5KVB3 A0A0P5MWG3 A0A0P5KSU2 A0A0P6FLY3 A0A023F2I8 A0A084WHT0 A0A0P5K323 A0A0P5T3I4 A0A0N8D582 A0A0P6FNM0 T1K1N8 A0A0P5UGB3 A0A067RIU1 A0A182T7Y8 A0A0P6G7S6 A0A0P5A418 A0A0P5TFB0 A0A0P5AL01 A0A0P5EHT2 W5J1Y7 A0A0P5ULQ1 A0A2M4AIB3 A0A224XEU1 A0A2M4BE31 A0A182GYU9 A0A0P5I8X5 A0A0P5R1Y0 E2BUL2
A0A2J7PR62 U5EYT8 A0A2J7PR47 A0A2J7PR48 A0A2J7PR63 A0A1Q3FPJ2 A0A1Q3FP78 A0A182R5C7 A0A1Q3FPU6 Q170L4 A0A2M4CJ21 A0A182UWY8 A0A182TS53 A0A182XNC5 A0A182JZY3 A0A182KSL1 A0A182PBS9 A0A182MYL4 A0A182QL14 A0A336LL51 A0A182HQ46 A0A182FU59 A0A182W944 A0A182M9X5 A0A182Y1R5 A0A0K8STY7 A0A0P5PPB9 A0A182JBS3 A0A0A9Z7L3 A0A0P5G5S6 A0A1L8E3B7 A0A1L8E3B6 A0A0P5H4F8 A0A0P5CIT8 T1JBH1 Q7Q082 A0A0P5Z9I4 A0A0N8CXC5 A0A0P5SZF3 A0A0P4ZG86 A0A0P5IU67 A0A0P6CTJ0 A0A0N8CEJ0 A0A0P5QQF6 A0A0P5WQD1 A0A0N8ARA9 A0A1B0EWS7 A0A0P5TWQ4 A0A0N8AJJ8 A0A0P6I4P9 A0A0P5SCA1 A0A0P6GS18 A0A0P5MPN1 A0A0N8BMI8 A0A0P5S9E5 A0A0P5E4B7 A0A0P5UAZ6 A0A0P6DCY0 A0A0P5EMH8 A0A0N8BPV7 A0A0P5XD67 B0WNK9 A0A0P6EAL7 A0A0P5EU30 A0A0P5KVB3 A0A0P5MWG3 A0A0P5KSU2 A0A0P6FLY3 A0A023F2I8 A0A084WHT0 A0A0P5K323 A0A0P5T3I4 A0A0N8D582 A0A0P6FNM0 T1K1N8 A0A0P5UGB3 A0A067RIU1 A0A182T7Y8 A0A0P6G7S6 A0A0P5A418 A0A0P5TFB0 A0A0P5AL01 A0A0P5EHT2 W5J1Y7 A0A0P5ULQ1 A0A2M4AIB3 A0A224XEU1 A0A2M4BE31 A0A182GYU9 A0A0P5I8X5 A0A0P5R1Y0 E2BUL2
PDB
3Q13
E-value=1.92889e-38,
Score=403
Ontologies
GO
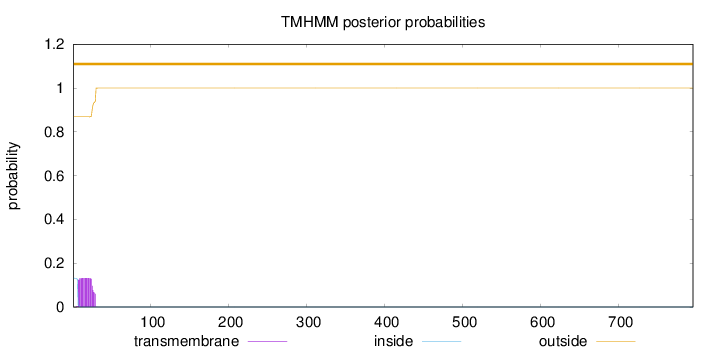
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.592879
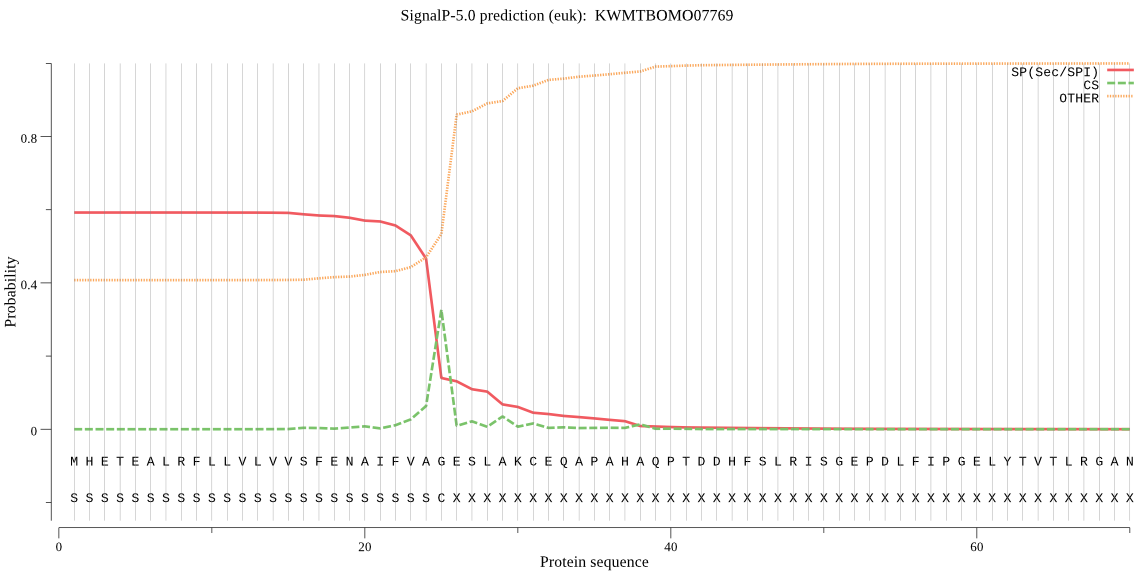
Length:
795
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.72416
Exp number, first 60 AAs:
2.72375
Total prob of N-in:
0.13165
outside
1 - 795
Population Genetic Test Statistics
Pi
215.195058
Theta
177.021515
Tajima's D
0.85202
CLR
0.315081
CSRT
0.619169041547923
Interpretation
Uncertain
