Gene
KWMTBOMO07768
Pre Gene Modal
BGIBMGA000896
Annotation
PREDICTED:_zinc_transporter_1_[Papilio_polytes]
Full name
Receptor protein-tyrosine kinase
Location in the cell
PlasmaMembrane Reliability : 4.111
Sequence
CDS
ATGGCAATGAAGGAATGGTTACAATGGCTGCCTCCACCACGATCACTATTAGCCCTCCTGTTGGCTGTCGCCGGCTTCGGCGGAAGATTATTCGCATCACACTTCACAAACTCTCCAACACTACTCGTTGACACGTGCCACTCTCTGTGTAGACTTGTGGGACTCATCACAACGCTTATTTCCTTTAAGTACGAAAGAGCAGACGAAGGAACCGGTCGCGAAGGTCGCCTGCGGAACACATTCGGATGGGCTCGCATCGAGGTCGTGGGTCGGCTATCTGTCCACGTGCTCTTCGCGTCTTTCGCTCTAGCGCTAGTGGTGAATGCTCTGCAACTTGGCGTGCATTCATCGCACGGGCCACCACCGCGCTACCCCAGAGTCATTGTCGGCAGCGCTGTAATAGGCTTGCTCTTTAACGCAATCAACTACATGCTACTTGCTGGTCGTGAACTCAGCTACAGTCGGCGACTGAGCGTCACTGAAGGTGGTGGTGTGGTGTTGAAGACTGGCTCAGGAGAACCAGTTCTGGCACACGCTCCTACTGATATCGCAAGCAGTCTGTTCGTGATAGCGGCTGGTCTCACAGAAGAGTGGGAACACGAGGCCTCTCGTATCGCGGACCCAGCCCTGTCTGCGTGTGCTGCTATCGTTCTCGTAGTATTCAACTATCCATTCATGCGTTCTGCTGGCCTTGTGCTCCTTCAAACCGTGCCCGAAGGTCTAGGAGCATGCGACTTGCGAACCGCCGCGCTCCGCGTCCCAGGAGTTTTCGCTATCCACGAACTGCACGTGTGGCAACTTCACAGGGATAAGGTGGTCGCCACGGCACACGTCATTTATGGAACGCACGAGGATTATCTCAAGAATTCAGCTCTAGTATGTGATGTTTTCAAACGCCACGGAATCGGGCTGGTCACTCTGCAGCCAGAATTCATGCTCACATCATCACGAGGTACGGAAGAAGAAAAGAAGGAATTAATAGCATATGGAAACTTAGGTTGTTCCTGTCCTTGCGTCAAAGAATGCACCGGTCCTCGTTGCTGCGAACCCCCAAACATACCACCTATTATACGAATTTAA
Protein
MAMKEWLQWLPPPRSLLALLLAVAGFGGRLFASHFTNSPTLLVDTCHSLCRLVGLITTLISFKYERADEGTGREGRLRNTFGWARIEVVGRLSVHVLFASFALALVVNALQLGVHSSHGPPPRYPRVIVGSAVIGLLFNAINYMLLAGRELSYSRRLSVTEGGGVVLKTGSGEPVLAHAPTDIASSLFVIAAGLTEEWEHEASRIADPALSACAAIVLVVFNYPFMRSAGLVLLQTVPEGLGACDLRTAALRVPGVFAIHELHVWQLHRDKVVATAHVIYGTHEDYLKNSALVCDVFKRHGIGLVTLQPEFMLTSSRGTEEEKKELIAYGNLGCSCPCVKECTGPRCCEPPNIPPIIRI
Summary
Catalytic Activity
ATP + L-tyrosyl-[protein] = ADP + H(+) + O-phospho-L-tyrosyl-[protein]
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A2H1V838
A0A2A4JI64
A0A2W1B3U7
H9IUG6
A0A194RSP5
A0A3S2NZC2
+ More
A0A194PJE7 A0A212FI09 A0A0L7LRV9 A0A1Y1KLC0 A0A0P6IY47 A0A1L8DEB2 A0A1W4W7B6 Q16SR1 A0A3B0J9T5 B3NIN2 J9JVW4 Q8SYV5 A0A2P8ZFU8 A0A026WEJ1 B4H7N3 Q29DC4 A0A336LZQ3 A0A3L8E0D5 B3M9B0 A0A2J7PKF2 Q9VPE3 Q7Q231 A0A182P1X1 B4IUG9 B4PET3 B4N6L6 A0A2H8TWK4 B4KY49 A0A2S2R0I2 A0A1W4W5R7 A0A232EUX5 E2AY53 A0A154PIC3 B4IAC0 A0A195BT85 A0A158P2D0 A0A2S2NTD9 A0A182YIX6 A0A182NV80 A0A182REI5 A0A310SDN7 A0A182SX84 A0A1B0CE91 A0A151WXM6 A0A182MC56 A0A195FHV6 A0A0L7REK6 A0A182W9J4 A0A195E548 A0A182QIP2 E2C777 K7IYU3 A0A182UK72 A0A182I5R5 A0A182KV99 A0A182FVV4 A0A067RV86 A0A182V522 W5JH22 A0A182JQU6 A0A3S3Q3Z0 U5EQ60 D2A0Y9 A0A087U9M7 A0A2J7PKF3 A0A2J7PKE6 A0A2J7PKG1 F4WUP2 A0A1B6FTW7 A0A226EP96 A0A1B6H5X6 N6TF74 A0A088AT30 A0A1B6IN37 U4UPH8 A0A2L2Y654 T1J125 E0VB97 A0A1J1HX04 A0A3R7LZE3 N6TQE0 A0A395I0F0 A0A017S738 A0A1L8DE99 A0A1D2N5R7 A0A2V5HKL2 A0A1L9RE43 A0A1L9VLN4 A0A1B6MAU5 E5SB51 A0A0V0RPM8 A0A0V1ADP1 A0A0V0TPK4 A0A0V0V4Z3
A0A194PJE7 A0A212FI09 A0A0L7LRV9 A0A1Y1KLC0 A0A0P6IY47 A0A1L8DEB2 A0A1W4W7B6 Q16SR1 A0A3B0J9T5 B3NIN2 J9JVW4 Q8SYV5 A0A2P8ZFU8 A0A026WEJ1 B4H7N3 Q29DC4 A0A336LZQ3 A0A3L8E0D5 B3M9B0 A0A2J7PKF2 Q9VPE3 Q7Q231 A0A182P1X1 B4IUG9 B4PET3 B4N6L6 A0A2H8TWK4 B4KY49 A0A2S2R0I2 A0A1W4W5R7 A0A232EUX5 E2AY53 A0A154PIC3 B4IAC0 A0A195BT85 A0A158P2D0 A0A2S2NTD9 A0A182YIX6 A0A182NV80 A0A182REI5 A0A310SDN7 A0A182SX84 A0A1B0CE91 A0A151WXM6 A0A182MC56 A0A195FHV6 A0A0L7REK6 A0A182W9J4 A0A195E548 A0A182QIP2 E2C777 K7IYU3 A0A182UK72 A0A182I5R5 A0A182KV99 A0A182FVV4 A0A067RV86 A0A182V522 W5JH22 A0A182JQU6 A0A3S3Q3Z0 U5EQ60 D2A0Y9 A0A087U9M7 A0A2J7PKF3 A0A2J7PKE6 A0A2J7PKG1 F4WUP2 A0A1B6FTW7 A0A226EP96 A0A1B6H5X6 N6TF74 A0A088AT30 A0A1B6IN37 U4UPH8 A0A2L2Y654 T1J125 E0VB97 A0A1J1HX04 A0A3R7LZE3 N6TQE0 A0A395I0F0 A0A017S738 A0A1L8DE99 A0A1D2N5R7 A0A2V5HKL2 A0A1L9RE43 A0A1L9VLN4 A0A1B6MAU5 E5SB51 A0A0V0RPM8 A0A0V1ADP1 A0A0V0TPK4 A0A0V0V4Z3
EC Number
2.7.10.1
Pubmed
28756777
19121390
26354079
22118469
26227816
28004739
+ More
26999592 17510324 17994087 29403074 24508170 15632085 30249741 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 17550304 28648823 20798317 21347285 25244985 20075255 20966253 24845553 20920257 23761445 18362917 19820115 21719571 23537049 26561354 20566863 24811710 27289101 28196534
26999592 17510324 17994087 29403074 24508170 15632085 30249741 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 17550304 28648823 20798317 21347285 25244985 20075255 20966253 24845553 20920257 23761445 18362917 19820115 21719571 23537049 26561354 20566863 24811710 27289101 28196534
EMBL
ODYU01000982
SOQ36562.1
NWSH01001481
PCG71120.1
KZ150481
PZC70728.1
+ More
BABH01001047 BABH01001048 BABH01001049 KQ459700 KPJ20370.1 RSAL01000084 RVE48405.1 KQ459606 KPI91210.1 AGBW02008449 OWR53356.1 JTDY01000230 KOB78167.1 GEZM01080351 GEZM01080350 JAV62203.1 GDUN01001132 JAN94787.1 GFDF01009359 JAV04725.1 CH477668 EAT37508.1 OUUW01000002 SPP77153.1 CH954178 EDV52528.2 ABLF02031677 ABLF02031680 AY071292 AAL48914.1 PYGN01000071 PSN55375.1 KK107250 EZA54363.1 CH479219 EDW34673.1 CH379070 EAL30490.1 UFQT01000226 SSX22053.1 QOIP01000002 RLU26181.1 CH902618 EDV40094.1 KPU78368.1 KPU78369.1 NEVH01024558 PNF16814.1 AE014296 BT150300 AAF51612.1 AAN12133.1 AGW24082.1 AAAB01008979 EAA13713.3 CH891906 EDX00033.1 KRK05359.1 KRK05360.1 CM000159 EDW95157.1 KRK02264.1 KRK02265.1 CH964161 EDW80005.1 GFXV01006868 MBW18673.1 CH933809 EDW18751.1 KRG06349.1 GGMS01014221 MBY83424.1 NNAY01002072 OXU22160.1 GL443762 EFN61649.1 KQ434900 KZC11068.1 CH480826 EDW44233.1 KQ976408 KYM91173.1 ADTU01007114 GGMR01007854 MBY20473.1 KQ773364 OAD52385.1 AJWK01008671 KQ982661 KYQ52662.1 AXCM01007287 KQ981523 KYN40275.1 KQ414609 KOC69260.1 KQ979657 KYN19979.1 AXCN02001805 GL453340 EFN76190.1 APCN01003487 KK852444 KDR23794.1 ADMH02001521 ETN62210.1 NCKU01000968 RWS13508.1 GANO01004395 JAB55476.1 KQ971338 EFA02578.1 KK118861 KFM74066.1 PNF16812.1 PNF16813.1 PNF16815.1 GL888375 EGI62039.1 GECZ01016119 JAS53650.1 LNIX01000002 OXA59463.1 GECU01037614 JAS70092.1 APGK01030223 APGK01030224 APGK01030225 APGK01030226 APGK01030227 KB740764 ENN79014.1 GECU01019375 JAS88331.1 KB632308 ERL91925.1 IAAA01013365 IAAA01013366 LAA03644.1 JH431752 DS235023 EEB10653.1 CVRI01000033 CRK92631.1 QCYY01003491 ROT63104.1 APGK01010575 APGK01010576 KB738221 ENN82719.1 KZ824278 RAL13672.1 KK088436 EYE92646.1 GFDF01009295 JAV04789.1 LJIJ01000196 ODN00619.1 KZ825102 PYI24291.1 KV878214 OJJ33191.1 KV878896 OJJ84836.1 GEBQ01006922 JAT33055.1 JYDH01000045 KRY36178.1 JYDL01000107 KRX16418.1 JYDQ01000006 KRY22955.1 JYDJ01000184 KRX40949.1 JYDN01000083 KRX58598.1
BABH01001047 BABH01001048 BABH01001049 KQ459700 KPJ20370.1 RSAL01000084 RVE48405.1 KQ459606 KPI91210.1 AGBW02008449 OWR53356.1 JTDY01000230 KOB78167.1 GEZM01080351 GEZM01080350 JAV62203.1 GDUN01001132 JAN94787.1 GFDF01009359 JAV04725.1 CH477668 EAT37508.1 OUUW01000002 SPP77153.1 CH954178 EDV52528.2 ABLF02031677 ABLF02031680 AY071292 AAL48914.1 PYGN01000071 PSN55375.1 KK107250 EZA54363.1 CH479219 EDW34673.1 CH379070 EAL30490.1 UFQT01000226 SSX22053.1 QOIP01000002 RLU26181.1 CH902618 EDV40094.1 KPU78368.1 KPU78369.1 NEVH01024558 PNF16814.1 AE014296 BT150300 AAF51612.1 AAN12133.1 AGW24082.1 AAAB01008979 EAA13713.3 CH891906 EDX00033.1 KRK05359.1 KRK05360.1 CM000159 EDW95157.1 KRK02264.1 KRK02265.1 CH964161 EDW80005.1 GFXV01006868 MBW18673.1 CH933809 EDW18751.1 KRG06349.1 GGMS01014221 MBY83424.1 NNAY01002072 OXU22160.1 GL443762 EFN61649.1 KQ434900 KZC11068.1 CH480826 EDW44233.1 KQ976408 KYM91173.1 ADTU01007114 GGMR01007854 MBY20473.1 KQ773364 OAD52385.1 AJWK01008671 KQ982661 KYQ52662.1 AXCM01007287 KQ981523 KYN40275.1 KQ414609 KOC69260.1 KQ979657 KYN19979.1 AXCN02001805 GL453340 EFN76190.1 APCN01003487 KK852444 KDR23794.1 ADMH02001521 ETN62210.1 NCKU01000968 RWS13508.1 GANO01004395 JAB55476.1 KQ971338 EFA02578.1 KK118861 KFM74066.1 PNF16812.1 PNF16813.1 PNF16815.1 GL888375 EGI62039.1 GECZ01016119 JAS53650.1 LNIX01000002 OXA59463.1 GECU01037614 JAS70092.1 APGK01030223 APGK01030224 APGK01030225 APGK01030226 APGK01030227 KB740764 ENN79014.1 GECU01019375 JAS88331.1 KB632308 ERL91925.1 IAAA01013365 IAAA01013366 LAA03644.1 JH431752 DS235023 EEB10653.1 CVRI01000033 CRK92631.1 QCYY01003491 ROT63104.1 APGK01010575 APGK01010576 KB738221 ENN82719.1 KZ824278 RAL13672.1 KK088436 EYE92646.1 GFDF01009295 JAV04789.1 LJIJ01000196 ODN00619.1 KZ825102 PYI24291.1 KV878214 OJJ33191.1 KV878896 OJJ84836.1 GEBQ01006922 JAT33055.1 JYDH01000045 KRY36178.1 JYDL01000107 KRX16418.1 JYDQ01000006 KRY22955.1 JYDJ01000184 KRX40949.1 JYDN01000083 KRX58598.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000283053
UP000053268
UP000007151
+ More
UP000037510 UP000192223 UP000008820 UP000268350 UP000008711 UP000007819 UP000245037 UP000053097 UP000008744 UP000001819 UP000279307 UP000007801 UP000235965 UP000000803 UP000007062 UP000075885 UP000002282 UP000007798 UP000009192 UP000192221 UP000215335 UP000000311 UP000076502 UP000001292 UP000078540 UP000005205 UP000076408 UP000075884 UP000075900 UP000075901 UP000092461 UP000075809 UP000075883 UP000078541 UP000053825 UP000075920 UP000078492 UP000075886 UP000008237 UP000002358 UP000075902 UP000075840 UP000075882 UP000069272 UP000027135 UP000075903 UP000000673 UP000075881 UP000285301 UP000007266 UP000054359 UP000007755 UP000198287 UP000019118 UP000005203 UP000030742 UP000009046 UP000183832 UP000283509 UP000248961 UP000019804 UP000094527 UP000249829 UP000184383 UP000184300 UP000054776 UP000054630 UP000054783 UP000055048 UP000054681
UP000037510 UP000192223 UP000008820 UP000268350 UP000008711 UP000007819 UP000245037 UP000053097 UP000008744 UP000001819 UP000279307 UP000007801 UP000235965 UP000000803 UP000007062 UP000075885 UP000002282 UP000007798 UP000009192 UP000192221 UP000215335 UP000000311 UP000076502 UP000001292 UP000078540 UP000005205 UP000076408 UP000075884 UP000075900 UP000075901 UP000092461 UP000075809 UP000075883 UP000078541 UP000053825 UP000075920 UP000078492 UP000075886 UP000008237 UP000002358 UP000075902 UP000075840 UP000075882 UP000069272 UP000027135 UP000075903 UP000000673 UP000075881 UP000285301 UP000007266 UP000054359 UP000007755 UP000198287 UP000019118 UP000005203 UP000030742 UP000009046 UP000183832 UP000283509 UP000248961 UP000019804 UP000094527 UP000249829 UP000184383 UP000184300 UP000054776 UP000054630 UP000054783 UP000055048 UP000054681
Interpro
IPR036837
Cation_efflux_CTD_sf
+ More
IPR002524 Cation_efflux
IPR027469 Cation_efflux_TMD_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR013098 Ig_I-set
IPR001824 Tyr_kinase_rcpt_3_CS
IPR011009 Kinase-like_dom_sf
IPR013783 Ig-like_fold
IPR013151 Immunoglobulin
IPR017441 Protein_kinase_ATP_BS
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR003598 Ig_sub2
IPR000719 Prot_kinase_dom
IPR002524 Cation_efflux
IPR027469 Cation_efflux_TMD_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR013098 Ig_I-set
IPR001824 Tyr_kinase_rcpt_3_CS
IPR011009 Kinase-like_dom_sf
IPR013783 Ig-like_fold
IPR013151 Immunoglobulin
IPR017441 Protein_kinase_ATP_BS
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR036179 Ig-like_dom_sf
IPR008266 Tyr_kinase_AS
IPR003598 Ig_sub2
IPR000719 Prot_kinase_dom
Gene 3D
ProteinModelPortal
A0A2H1V838
A0A2A4JI64
A0A2W1B3U7
H9IUG6
A0A194RSP5
A0A3S2NZC2
+ More
A0A194PJE7 A0A212FI09 A0A0L7LRV9 A0A1Y1KLC0 A0A0P6IY47 A0A1L8DEB2 A0A1W4W7B6 Q16SR1 A0A3B0J9T5 B3NIN2 J9JVW4 Q8SYV5 A0A2P8ZFU8 A0A026WEJ1 B4H7N3 Q29DC4 A0A336LZQ3 A0A3L8E0D5 B3M9B0 A0A2J7PKF2 Q9VPE3 Q7Q231 A0A182P1X1 B4IUG9 B4PET3 B4N6L6 A0A2H8TWK4 B4KY49 A0A2S2R0I2 A0A1W4W5R7 A0A232EUX5 E2AY53 A0A154PIC3 B4IAC0 A0A195BT85 A0A158P2D0 A0A2S2NTD9 A0A182YIX6 A0A182NV80 A0A182REI5 A0A310SDN7 A0A182SX84 A0A1B0CE91 A0A151WXM6 A0A182MC56 A0A195FHV6 A0A0L7REK6 A0A182W9J4 A0A195E548 A0A182QIP2 E2C777 K7IYU3 A0A182UK72 A0A182I5R5 A0A182KV99 A0A182FVV4 A0A067RV86 A0A182V522 W5JH22 A0A182JQU6 A0A3S3Q3Z0 U5EQ60 D2A0Y9 A0A087U9M7 A0A2J7PKF3 A0A2J7PKE6 A0A2J7PKG1 F4WUP2 A0A1B6FTW7 A0A226EP96 A0A1B6H5X6 N6TF74 A0A088AT30 A0A1B6IN37 U4UPH8 A0A2L2Y654 T1J125 E0VB97 A0A1J1HX04 A0A3R7LZE3 N6TQE0 A0A395I0F0 A0A017S738 A0A1L8DE99 A0A1D2N5R7 A0A2V5HKL2 A0A1L9RE43 A0A1L9VLN4 A0A1B6MAU5 E5SB51 A0A0V0RPM8 A0A0V1ADP1 A0A0V0TPK4 A0A0V0V4Z3
A0A194PJE7 A0A212FI09 A0A0L7LRV9 A0A1Y1KLC0 A0A0P6IY47 A0A1L8DEB2 A0A1W4W7B6 Q16SR1 A0A3B0J9T5 B3NIN2 J9JVW4 Q8SYV5 A0A2P8ZFU8 A0A026WEJ1 B4H7N3 Q29DC4 A0A336LZQ3 A0A3L8E0D5 B3M9B0 A0A2J7PKF2 Q9VPE3 Q7Q231 A0A182P1X1 B4IUG9 B4PET3 B4N6L6 A0A2H8TWK4 B4KY49 A0A2S2R0I2 A0A1W4W5R7 A0A232EUX5 E2AY53 A0A154PIC3 B4IAC0 A0A195BT85 A0A158P2D0 A0A2S2NTD9 A0A182YIX6 A0A182NV80 A0A182REI5 A0A310SDN7 A0A182SX84 A0A1B0CE91 A0A151WXM6 A0A182MC56 A0A195FHV6 A0A0L7REK6 A0A182W9J4 A0A195E548 A0A182QIP2 E2C777 K7IYU3 A0A182UK72 A0A182I5R5 A0A182KV99 A0A182FVV4 A0A067RV86 A0A182V522 W5JH22 A0A182JQU6 A0A3S3Q3Z0 U5EQ60 D2A0Y9 A0A087U9M7 A0A2J7PKF3 A0A2J7PKE6 A0A2J7PKG1 F4WUP2 A0A1B6FTW7 A0A226EP96 A0A1B6H5X6 N6TF74 A0A088AT30 A0A1B6IN37 U4UPH8 A0A2L2Y654 T1J125 E0VB97 A0A1J1HX04 A0A3R7LZE3 N6TQE0 A0A395I0F0 A0A017S738 A0A1L8DE99 A0A1D2N5R7 A0A2V5HKL2 A0A1L9RE43 A0A1L9VLN4 A0A1B6MAU5 E5SB51 A0A0V0RPM8 A0A0V1ADP1 A0A0V0TPK4 A0A0V0V4Z3
Ontologies
GO
Topology
SignalP
Position: 1 - 32,
Likelihood: 0.612730
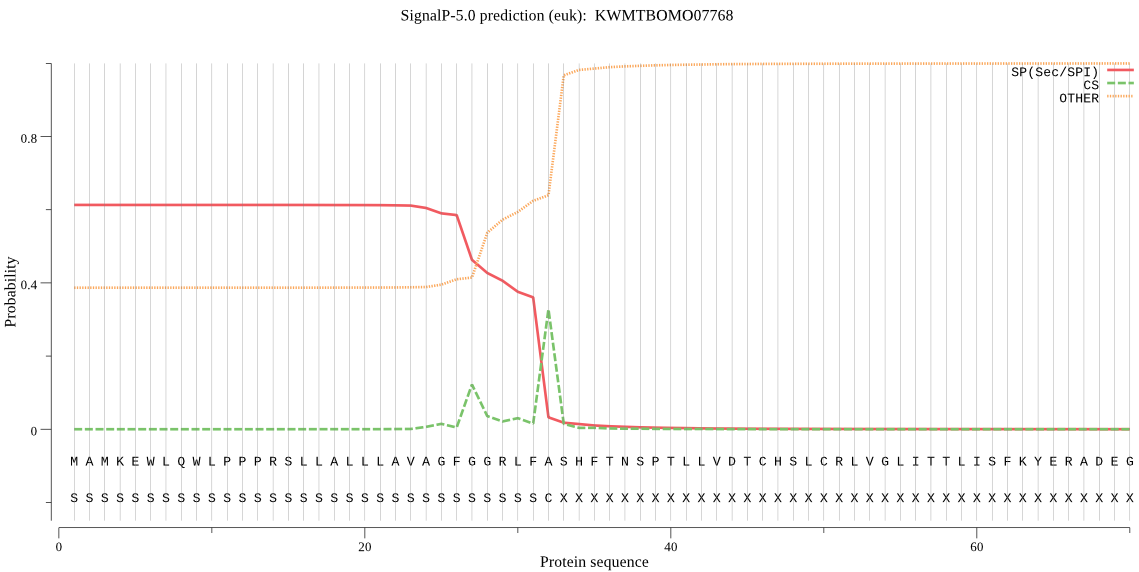
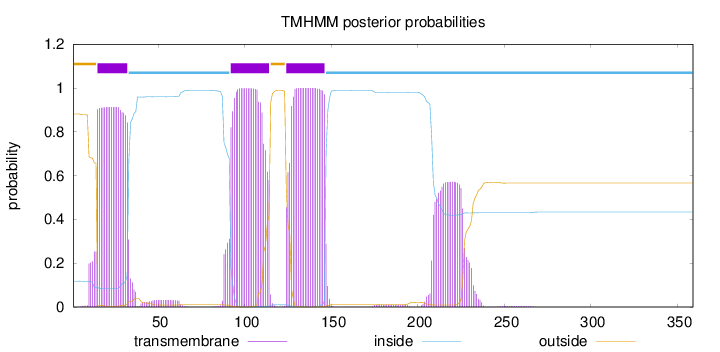
Length:
359
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
74.8043900000001
Exp number, first 60 AAs:
18.67261
Total prob of N-in:
0.11806
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 32
inside
33 - 91
TMhelix
92 - 114
outside
115 - 123
TMhelix
124 - 146
inside
147 - 359
Population Genetic Test Statistics
Pi
178.587766
Theta
153.692574
Tajima's D
0.744385
CLR
0.088386
CSRT
0.587620618969052
Interpretation
Uncertain
