Gene
KWMTBOMO07756
Pre Gene Modal
BGIBMGA000891
Annotation
ionotropic_GABA_receptor_subunit_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.39
Sequence
CDS
ATGTTGACATCCCATGACGAAGCTACGCCGCGGCACACTCCAAAGATTTTGTCTGCGGCGAAAAACACGTCACGAGATACCGCAGAACCCGACTACTGGGACAACATAAATGAAATAATGTTTTCCAATTACGCCAACTCTAGTTTTGAACAGACAGACGTGTTTACGTGGAAACAGAACAAGAGAAGTATAAACGATGCCGTCTCTAAAAATATAACTTCTGTTTTGGAGAACCTTTTGAAGAACTATGAAAATTCTCAGTTGCCAACGCATGGTAAAGGTTATCCAACTGTAGTTCAAACCAACATCCTAATAAGGAGCATGGGACCAGTTTCAGAACTCGATATGGATTATTCGATGGATTGCTACTTCAGGCAATATTGGCGTGACACGAGGCTGTCGTTTCTCGGTCCCATCCGCTCTTTGTCCCTTTCCATCAAGATGCTCGAGAGGATTTGGAGGCCAGACACGTATTTTTACAACGGAAAACATTCCTATGTACACACTATCACTGTACCGAATAAACTGCTTCGCATCAGTCAACACGGTGATATTTTGTATTCAATGAGATTAACTATTAAAGCTAAATGCCCTATGGAACTTCGTAATTTTCCGATGGATCGTCAATCGTGCCCTCTTATCCTTGGCAGCTACGCCTACTCGAATCAACAACTAGTTTATCAATGGCAAAACTCGCAGAGCGTCAACTTTGTTCCCGGAATGACCCTCTCACAGTTTGACCTTATCAGTTTTCCGTACAGAAATTTCACATTTACCAGACGAGAAGGTGATTTTTCAGTGTTACAAGTCTCTTTCAACCTCAAAAGGCACACCGGCTATTTTCTTATTCAAGTGTACGTGCCCTGCATCCTTATAGTAGTCCTTTCTTGGGTCTCGTTCTGGATTCATCGCGAGGCTACATCCGATCGCGTGGGCTTAGGTATTACCACTGTGCTAACTCTGTCCACGATAAGTCTTGATTCTCGTACCGATTTGCCTAAAGTTCGCTACGCAACCGCACTGGATTGGTTCTTGCTAATGAGTTTCTTTTACTGCATCGCAACTCTACTAGAATTCGCTGGAGTGCATTATTTCACGAAGGTCGGTTCTGGTGAAATAGTTATCGACGATTCGGAATGGGAAGAGCTGATCGAAGAAGTCGGAGGGGACGCGGTCGCGGCACGTGAACTCGCCGTACGTCGTCGTAGTTCTGCTAGATCCGCCACTAATAACCTCTCCATGCCATCCCAAAGTTCAAACCCAACGACTTCGGATAACTTGGAAACTGAACAGCCAGTTGCAGTCCGCCTCACCATGGAACGTACAACTCAGACCGAGACCAGAGTGCCTCGTTGGAGACAACTCTTGTACTGCTTAGCTGGCGACGATCGCTACAGACGACAGAGACAACGGGAAGCAGGGAGTAGGGGACACATCAACAGCGTCTCACACATCGACAGGGCTGCACGTGTTTTATTCCCGGCTTCGTTTGCACTGCTCAATCTATTCTACTGGATGATATATGCCTTCTCGAGTGATGACTTTGCCTGGAGCGACAATCCCATTAATACCTTATCGCATTGA
Protein
MLTSHDEATPRHTPKILSAAKNTSRDTAEPDYWDNINEIMFSNYANSSFEQTDVFTWKQNKRSINDAVSKNITSVLENLLKNYENSQLPTHGKGYPTVVQTNILIRSMGPVSELDMDYSMDCYFRQYWRDTRLSFLGPIRSLSLSIKMLERIWRPDTYFYNGKHSYVHTITVPNKLLRISQHGDILYSMRLTIKAKCPMELRNFPMDRQSCPLILGSYAYSNQQLVYQWQNSQSVNFVPGMTLSQFDLISFPYRNFTFTRREGDFSVLQVSFNLKRHTGYFLIQVYVPCILIVVLSWVSFWIHREATSDRVGLGITTVLTLSTISLDSRTDLPKVRYATALDWFLLMSFFYCIATLLEFAGVHYFTKVGSGEIVIDDSEWEELIEEVGGDAVAARELAVRRRSSARSATNNLSMPSQSSNPTTSDNLETEQPVAVRLTMERTTQTETRVPRWRQLLYCLAGDDRYRRQRQREAGSRGHINSVSHIDRAARVLFPASFALLNLFYWMIYAFSSDDFAWSDNPINTLSH
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
A0A0U4VGB7
E0X9H5
A0A2W1BKQ8
A0A2A4J4D2
A0A194RSQ9
A0A194PDV1
+ More
A0A3G1J4Z0 H9IUG1 A0A2H1VYI0 D2A157 A8DMV0 A0A1Y1MAX8 A0A139WJI2 A0A195C9D5 A0A067RK81 A0A158NC41 A0A154PBZ7 A0A195FL08 A0A0C9QIW0 E1ZY67 A0A2J7RFP2 E9J9I1 A0A3L8DWG4 A0A1C9HM80 A0A026VY51 A0A0L7R7A6 A0A182V204 A0A182HIT2 A0A182X2D2 A0A182TTZ4 A0A182TAD2 A0A182M2G4 A0A182XYF7 A0A182IXG1 A0A182N0V1 A0A182VVK4 A0A084VA97 A0NCM7 A0A182R202 A0A182QI39 A0A146MBU7 W5JRF7 N6UNK1 A0A336LLT9 Q0GQQ6 A0A182FK43 A0A232FFQ0 B4L420 A0A0L7L8T8 B4JLF8 B5DSR3 A0A1B0GIH3 B4NPE7 B4M3E7 A0A3Q0JAE5 A0A1B0DRB5 A0A3Q0JAF5 Q9VXL9 B3NXH5 B4PX93 A0A1A9YM90 A0A1A9VSX5 E1JJA9 U4UPN7 A0A1I8MM43 A0A1W4V2W7 A0A1B0BT40 B4IKF9 A0A182PNS5 A0A1J1IXM4 A0A0L0BLK0 A0A0M8ZRL4 A0A1B0G1E6 A0A1A9WIN9 B4HCI5 A0A182K9S2 A0A226E691 N6UNC3 N6TVP1 A0A088A2R6 A0A1B0A1T7 A0A212FHY0 R9S1I4 Q4V4W7 R7UPT2
A0A3G1J4Z0 H9IUG1 A0A2H1VYI0 D2A157 A8DMV0 A0A1Y1MAX8 A0A139WJI2 A0A195C9D5 A0A067RK81 A0A158NC41 A0A154PBZ7 A0A195FL08 A0A0C9QIW0 E1ZY67 A0A2J7RFP2 E9J9I1 A0A3L8DWG4 A0A1C9HM80 A0A026VY51 A0A0L7R7A6 A0A182V204 A0A182HIT2 A0A182X2D2 A0A182TTZ4 A0A182TAD2 A0A182M2G4 A0A182XYF7 A0A182IXG1 A0A182N0V1 A0A182VVK4 A0A084VA97 A0NCM7 A0A182R202 A0A182QI39 A0A146MBU7 W5JRF7 N6UNK1 A0A336LLT9 Q0GQQ6 A0A182FK43 A0A232FFQ0 B4L420 A0A0L7L8T8 B4JLF8 B5DSR3 A0A1B0GIH3 B4NPE7 B4M3E7 A0A3Q0JAE5 A0A1B0DRB5 A0A3Q0JAF5 Q9VXL9 B3NXH5 B4PX93 A0A1A9YM90 A0A1A9VSX5 E1JJA9 U4UPN7 A0A1I8MM43 A0A1W4V2W7 A0A1B0BT40 B4IKF9 A0A182PNS5 A0A1J1IXM4 A0A0L0BLK0 A0A0M8ZRL4 A0A1B0G1E6 A0A1A9WIN9 B4HCI5 A0A182K9S2 A0A226E691 N6UNC3 N6TVP1 A0A088A2R6 A0A1B0A1T7 A0A212FHY0 R9S1I4 Q4V4W7 R7UPT2
Pubmed
20924418
28756777
26354079
19121390
18362917
19820115
+ More
17880682 28004739 24845553 21347285 20798317 21282665 30249741 24508170 25244985 24438588 12364791 26823975 20920257 23761445 23537049 16902773 28648823 17994087 26227816 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 26108605 22118469 23254933
17880682 28004739 24845553 21347285 20798317 21282665 30249741 24508170 25244985 24438588 12364791 26823975 20920257 23761445 23537049 16902773 28648823 17994087 26227816 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25315136 26108605 22118469 23254933
EMBL
AB278157
BAT57342.1
GQ890659
ADM88004.1
KZ149987
PZC75662.1
+ More
NWSH01003070 PCG67007.1 KQ459700 KPJ20385.1 KQ459606 KPI91198.1 KX856967 ASY91959.1 BABH01001068 ODYU01004917 SOQ45284.1 KQ971338 EFA02907.2 EF545127 ABU63605.1 GEZM01038969 JAV81465.1 KYB28119.1 KQ978068 KYM97474.1 KK852422 KDR24266.1 ADTU01011491 ADTU01011492 ADTU01011493 KQ434869 KZC09372.1 KQ981491 KYN41031.1 GBYB01014668 JAG84435.1 GL435171 EFN73916.1 NEVH01004410 PNF39640.1 GL769313 EFZ10522.1 QOIP01000004 RLU24118.1 KX355314 AOO87784.1 KK107785 EZA47794.1 KQ414642 KOC66724.1 APCN01004087 AXCM01014059 AXCP01007153 ATLV01003051 KE524109 KFB34891.1 AAAB01008846 EAU77351.2 AXCN02001228 GDHC01002117 JAQ16512.1 ADMH02000632 ETN65490.1 APGK01004349 KB735711 ENN83325.1 UFQT01000005 SSX17327.1 DQ667193 ABG75745.1 NNAY01000335 OXU29127.1 CH933810 EDW07298.1 JTDY01002176 KOB71948.1 CH916370 EDW00411.1 CH475585 EDY70824.2 AJWK01014118 CH964291 EDW86387.2 CH940651 EDW65322.2 AJVK01019838 AE014298 BT022901 AAF48539.4 AAY55317.1 CH954180 EDV46934.2 CM000162 EDX01856.2 ACZ95305.1 KB632308 ERL91985.1 JXJN01019977 CH480852 EDW51551.1 CVRI01000063 CRL04832.1 JRES01001701 KNC20828.1 KQ435951 KOX68119.1 CCAG010015963 CH479336 EDW26951.1 LNIX01000006 OXA53155.1 APGK01005252 KB736144 ENN83250.1 APGK01016425 APGK01016426 KB739845 ENN82103.1 AGBW02008449 OWR53344.1 KC710947 AGN03795.1 BT022889 AAY55305.1 AMQN01006847 KB299334 ELU08088.1
NWSH01003070 PCG67007.1 KQ459700 KPJ20385.1 KQ459606 KPI91198.1 KX856967 ASY91959.1 BABH01001068 ODYU01004917 SOQ45284.1 KQ971338 EFA02907.2 EF545127 ABU63605.1 GEZM01038969 JAV81465.1 KYB28119.1 KQ978068 KYM97474.1 KK852422 KDR24266.1 ADTU01011491 ADTU01011492 ADTU01011493 KQ434869 KZC09372.1 KQ981491 KYN41031.1 GBYB01014668 JAG84435.1 GL435171 EFN73916.1 NEVH01004410 PNF39640.1 GL769313 EFZ10522.1 QOIP01000004 RLU24118.1 KX355314 AOO87784.1 KK107785 EZA47794.1 KQ414642 KOC66724.1 APCN01004087 AXCM01014059 AXCP01007153 ATLV01003051 KE524109 KFB34891.1 AAAB01008846 EAU77351.2 AXCN02001228 GDHC01002117 JAQ16512.1 ADMH02000632 ETN65490.1 APGK01004349 KB735711 ENN83325.1 UFQT01000005 SSX17327.1 DQ667193 ABG75745.1 NNAY01000335 OXU29127.1 CH933810 EDW07298.1 JTDY01002176 KOB71948.1 CH916370 EDW00411.1 CH475585 EDY70824.2 AJWK01014118 CH964291 EDW86387.2 CH940651 EDW65322.2 AJVK01019838 AE014298 BT022901 AAF48539.4 AAY55317.1 CH954180 EDV46934.2 CM000162 EDX01856.2 ACZ95305.1 KB632308 ERL91985.1 JXJN01019977 CH480852 EDW51551.1 CVRI01000063 CRL04832.1 JRES01001701 KNC20828.1 KQ435951 KOX68119.1 CCAG010015963 CH479336 EDW26951.1 LNIX01000006 OXA53155.1 APGK01005252 KB736144 ENN83250.1 APGK01016425 APGK01016426 KB739845 ENN82103.1 AGBW02008449 OWR53344.1 KC710947 AGN03795.1 BT022889 AAY55305.1 AMQN01006847 KB299334 ELU08088.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000005204
UP000007266
UP000078542
+ More
UP000027135 UP000005205 UP000076502 UP000078541 UP000000311 UP000235965 UP000279307 UP000053097 UP000053825 UP000075903 UP000075840 UP000076407 UP000075902 UP000075901 UP000075883 UP000076408 UP000075880 UP000075884 UP000075920 UP000030765 UP000007062 UP000075900 UP000075886 UP000000673 UP000019118 UP000069272 UP000215335 UP000009192 UP000037510 UP000001070 UP000001819 UP000092461 UP000007798 UP000008792 UP000079169 UP000092462 UP000000803 UP000008711 UP000002282 UP000092443 UP000078200 UP000030742 UP000095301 UP000192221 UP000092460 UP000001292 UP000075885 UP000183832 UP000037069 UP000053105 UP000092444 UP000091820 UP000008744 UP000075881 UP000198287 UP000005203 UP000092445 UP000007151 UP000014760
UP000027135 UP000005205 UP000076502 UP000078541 UP000000311 UP000235965 UP000279307 UP000053097 UP000053825 UP000075903 UP000075840 UP000076407 UP000075902 UP000075901 UP000075883 UP000076408 UP000075880 UP000075884 UP000075920 UP000030765 UP000007062 UP000075900 UP000075886 UP000000673 UP000019118 UP000069272 UP000215335 UP000009192 UP000037510 UP000001070 UP000001819 UP000092461 UP000007798 UP000008792 UP000079169 UP000092462 UP000000803 UP000008711 UP000002282 UP000092443 UP000078200 UP000030742 UP000095301 UP000192221 UP000092460 UP000001292 UP000075885 UP000183832 UP000037069 UP000053105 UP000092444 UP000091820 UP000008744 UP000075881 UP000198287 UP000005203 UP000092445 UP000007151 UP000014760
Interpro
Gene 3D
ProteinModelPortal
A0A0U4VGB7
E0X9H5
A0A2W1BKQ8
A0A2A4J4D2
A0A194RSQ9
A0A194PDV1
+ More
A0A3G1J4Z0 H9IUG1 A0A2H1VYI0 D2A157 A8DMV0 A0A1Y1MAX8 A0A139WJI2 A0A195C9D5 A0A067RK81 A0A158NC41 A0A154PBZ7 A0A195FL08 A0A0C9QIW0 E1ZY67 A0A2J7RFP2 E9J9I1 A0A3L8DWG4 A0A1C9HM80 A0A026VY51 A0A0L7R7A6 A0A182V204 A0A182HIT2 A0A182X2D2 A0A182TTZ4 A0A182TAD2 A0A182M2G4 A0A182XYF7 A0A182IXG1 A0A182N0V1 A0A182VVK4 A0A084VA97 A0NCM7 A0A182R202 A0A182QI39 A0A146MBU7 W5JRF7 N6UNK1 A0A336LLT9 Q0GQQ6 A0A182FK43 A0A232FFQ0 B4L420 A0A0L7L8T8 B4JLF8 B5DSR3 A0A1B0GIH3 B4NPE7 B4M3E7 A0A3Q0JAE5 A0A1B0DRB5 A0A3Q0JAF5 Q9VXL9 B3NXH5 B4PX93 A0A1A9YM90 A0A1A9VSX5 E1JJA9 U4UPN7 A0A1I8MM43 A0A1W4V2W7 A0A1B0BT40 B4IKF9 A0A182PNS5 A0A1J1IXM4 A0A0L0BLK0 A0A0M8ZRL4 A0A1B0G1E6 A0A1A9WIN9 B4HCI5 A0A182K9S2 A0A226E691 N6UNC3 N6TVP1 A0A088A2R6 A0A1B0A1T7 A0A212FHY0 R9S1I4 Q4V4W7 R7UPT2
A0A3G1J4Z0 H9IUG1 A0A2H1VYI0 D2A157 A8DMV0 A0A1Y1MAX8 A0A139WJI2 A0A195C9D5 A0A067RK81 A0A158NC41 A0A154PBZ7 A0A195FL08 A0A0C9QIW0 E1ZY67 A0A2J7RFP2 E9J9I1 A0A3L8DWG4 A0A1C9HM80 A0A026VY51 A0A0L7R7A6 A0A182V204 A0A182HIT2 A0A182X2D2 A0A182TTZ4 A0A182TAD2 A0A182M2G4 A0A182XYF7 A0A182IXG1 A0A182N0V1 A0A182VVK4 A0A084VA97 A0NCM7 A0A182R202 A0A182QI39 A0A146MBU7 W5JRF7 N6UNK1 A0A336LLT9 Q0GQQ6 A0A182FK43 A0A232FFQ0 B4L420 A0A0L7L8T8 B4JLF8 B5DSR3 A0A1B0GIH3 B4NPE7 B4M3E7 A0A3Q0JAE5 A0A1B0DRB5 A0A3Q0JAF5 Q9VXL9 B3NXH5 B4PX93 A0A1A9YM90 A0A1A9VSX5 E1JJA9 U4UPN7 A0A1I8MM43 A0A1W4V2W7 A0A1B0BT40 B4IKF9 A0A182PNS5 A0A1J1IXM4 A0A0L0BLK0 A0A0M8ZRL4 A0A1B0G1E6 A0A1A9WIN9 B4HCI5 A0A182K9S2 A0A226E691 N6UNC3 N6TVP1 A0A088A2R6 A0A1B0A1T7 A0A212FHY0 R9S1I4 Q4V4W7 R7UPT2
PDB
6HUG
E-value=8.09788e-78,
Score=740
Ontologies
GO
PANTHER
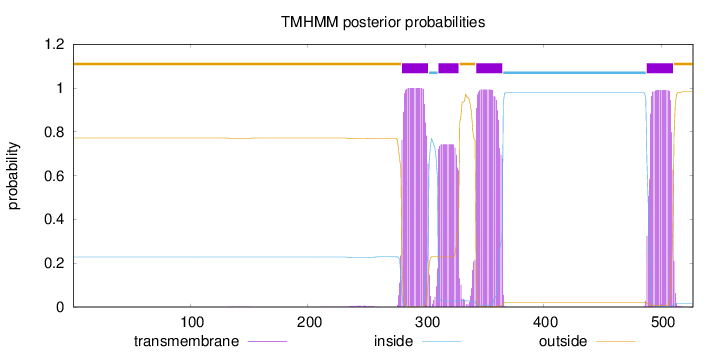
Topology
Length:
527
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
79.4107400000001
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.22863
outside
1 - 279
TMhelix
280 - 302
inside
303 - 310
TMhelix
311 - 328
outside
329 - 342
TMhelix
343 - 365
inside
366 - 487
TMhelix
488 - 510
outside
511 - 527
Population Genetic Test Statistics
Pi
264.442158
Theta
180.158876
Tajima's D
1.253777
CLR
0.030074
CSRT
0.726613669316534
Interpretation
Uncertain
