Gene
KWMTBOMO07750
Pre Gene Modal
BGIBMGA000890
Annotation
PREDICTED:_sorting_nexin-25_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.077 Nuclear Reliability : 1.739
Sequence
CDS
ATGTTGTGGATGTTAATATGTGGTGTTCCACTAGTTTACGTTTTGATTTGGTATTTTCCAATATTATGGAGCCTAATTTATTTGTTATTGTTAGCTATCTTTATAATATTAAGTGTATTCACTCTTAGTTTATGGGGGCATATTGCATTGTCATCACCTTATCATATACCACCGTTTTTGATAGAAAGAATTGAAAAGGAAGTTCGACAATTTGAAGAAGCACTGAAGGAAGATCAGGCAAATTGGCCTCAATCCATGAAAAAATCTCATCTGCCAGTCATATTCGGAAGAACTGTAGACAGTCAATTGCAGCTTCTCATTGATTACATATTGAGAGATTTTGTAACTCGATGGCTGAAAGAATTGGCACATAAACCTGAACCAGTCATAGACAAATTCAAGGAGCATATTTGGGGAGGTATACAAAATTTATATGAAAGGCTATCAAGGGTTGATGCTGAAAAATTACTGGCTAATGATATGGTGGTGAAGATAACTCAACATTTTGAAAGGATAAGAATTGCAAGAAGTTGTGCACTAGAATTGAATCAAGCACCTGTTTTTGCTCTAGCACCCCATCTGACATCTTTCGAAACAGAACAGCATTATTTGAGGCAGATCAGTGAACTTATTATCATGTTTTTGTTGCCGAGATGCTATTCTTTGGCACCAGGATGCCATCTCATGAGGGAAATACTTGCCTGCAAGATTTTACAACCAGCTATAGATCTTGTAACAGAACCAGATTATATAAATCAGAAAATTATTCAATATTTGGAAGCACAAAAAGAAGTAAACGCCATGCATATAAGGACTCATGAATATGCAAAAACGTTTGAGGACTACATACGTCTCATTAATAGTTGCAACAATGTTGACACGCTGAAACGGTTACGGTATGATATAGTAACACAAATAATGCAAGCAACAACGCTCCAAAACGTGAAACGAGCAAAAGGAATTGACCTAGATGCAATAGAAAAAAGTAATACACAACACAACATAAGTCGGCAACAAATAACCGATGCTAAGAAACTTCAGAGGTATATAAATCAACTTACAATAGCGAAAGCGGAGTGTGAAACTGCACTTAGGAAAATGGGCTGGGATGGTGCGTTTCCTACATTTGAGACAGATAGCAAGACGCTACCTTTGCACAAAGTCATGGACAGCGTGACGGGGCGCCGGTATTTGTCGATGTTCTTAGAAAAAATGTGTTCTCAAGGCCTAGTTGGCTTTTGGACGTCAGTGGAAGACCTGCGGCATAGCCCCAGGAGCAGCTGGCATCAGCTCGGTGCCGAGATATTTTATACTTACATAAGATCGCCTAGTGCGGAAATTAAAGTCGATAAGGACACTAGAAAGCGAATGGAATCCTTTCTGCTGGGCGACAAGGGACCAGATGTATTTTATGAAGTACAAGAGACCGTTCTTGATACAATACAAGACAAATATTATCATTCGTTCTTAATGAGTGAGGAATATAAAGCTTTAATTGCTGAACTTGCAACAGAAGAAAATAGCAAAGATACGAACGTGGATAGATCGCCAATAGAAGAGCGGCAATTGTCGAATGAGTCTGTTTCTTCGGCGGAGAGCGTCGCGGGAGCGTTGCACCTAACGGAGCACTCGACGTACGCACGAAGAAAACTGGATCAGCTACAAGAACGACACAACAATAAGATACAGGCTCTAGCTGCTCTGCGAGCGTCGCTGAAGCCGGAGTCCCCTGCCTTGTCCATGCTGGCCAACGAGGTGGAGAAGTTGGCCGGTGAACAGATGAGGCTCGAGGCACACCTGGCCAGGACGGACACGTGGGCCGAGCACTTGGGGCGCTGGCGGGCCTCCGTCCACAGTGCTGACGTAATTGACGAGTCTAAACCACCGCAGTTTGTGATAGTCGTGCACATGGCGGAAAAGGAAGATTCGAACAGCGGGGACCGACCCGAGCAGATCACCACCGGCTGGGTCCTGTTGAGGACGCTCAATGAATTCCAGGACTTACATCGAAAGTTAAGGCCTTTGTGTTCAGAATTGAAAAATTTAGAGCTACCCTCAAACTCTTTTAAATTTTTATTCGGAAAAAACGATAAGAATTCTTTAGACAAAGCAAAAACATTAATACAAAAGTATTTAGAGTTCGTATTAGAAGACGACAGACTAAACCAAAGTGAAGCTTTATATACATTCCTAAACCCTAGCTCGGAGTATTTAAAACAAGAGGACATGCCAAAGAAAAATAAATTCTCATTATCTACATTATTCAAAAGCGCAAGCAGCGAAGCAACGAATAGATCGTCGCAGGAGAAGGAGGCCTTTAACCACATGTCGGCGGACGAAGACGAAATATCGATGTACCTGGATGGCAACGGAGCCGAGGCGGGAAACAAAACGATGACTAATTCCTTGCGAGGGGCCAGCGCGATAGCCGAAGAACGGGACAGCACCGCGGAGCCGCTGTACGCGCTGCTCAGCGAGGTGTTCGACATGCGTGGCGTGTTCCGCTGGCTCCGGAAGACGCTCGTCACCTTCGTGCAGATCACGTACGGACGCACCATCAATAGACAAATAAAAGATACAATATCTTGGCTGTTTTCGGAGCAGATGTTACATTATTACATAAGCGTAGTTATCAAGTCTTGGTGGCCGGGCGGAGTCCTCTCGGAGCACGGCTCGAACAGGAATTTTCAAGATAAAGAGCACACGAGGACCCTCGCGCTGCAGCAGCTGAGCGAGAGCGTGATGGACGCGCTGTCGTCGCTGGTGGGCGCGCAGACGGCGGCCCGCGGCGCGCACAAGCTGTTCCACACGCTGCAGCACACCACGCACAACAAACAACTCTTCTACGAATTGTTCGAACTGGTATTGATAGAAGTATTTCCTGAGCTCAAGCGTTACTATAACTAA
Protein
MLWMLICGVPLVYVLIWYFPILWSLIYLLLLAIFIILSVFTLSLWGHIALSSPYHIPPFLIERIEKEVRQFEEALKEDQANWPQSMKKSHLPVIFGRTVDSQLQLLIDYILRDFVTRWLKELAHKPEPVIDKFKEHIWGGIQNLYERLSRVDAEKLLANDMVVKITQHFERIRIARSCALELNQAPVFALAPHLTSFETEQHYLRQISELIIMFLLPRCYSLAPGCHLMREILACKILQPAIDLVTEPDYINQKIIQYLEAQKEVNAMHIRTHEYAKTFEDYIRLINSCNNVDTLKRLRYDIVTQIMQATTLQNVKRAKGIDLDAIEKSNTQHNISRQQITDAKKLQRYINQLTIAKAECETALRKMGWDGAFPTFETDSKTLPLHKVMDSVTGRRYLSMFLEKMCSQGLVGFWTSVEDLRHSPRSSWHQLGAEIFYTYIRSPSAEIKVDKDTRKRMESFLLGDKGPDVFYEVQETVLDTIQDKYYHSFLMSEEYKALIAELATEENSKDTNVDRSPIEERQLSNESVSSAESVAGALHLTEHSTYARRKLDQLQERHNNKIQALAALRASLKPESPALSMLANEVEKLAGEQMRLEAHLARTDTWAEHLGRWRASVHSADVIDESKPPQFVIVVHMAEKEDSNSGDRPEQITTGWVLLRTLNEFQDLHRKLRPLCSELKNLELPSNSFKFLFGKNDKNSLDKAKTLIQKYLEFVLEDDRLNQSEALYTFLNPSSEYLKQEDMPKKNKFSLSTLFKSASSEATNRSSQEKEAFNHMSADEDEISMYLDGNGAEAGNKTMTNSLRGASAIAEERDSTAEPLYALLSEVFDMRGVFRWLRKTLVTFVQITYGRTINRQIKDTISWLFSEQMLHYYISVVIKSWWPGGVLSEHGSNRNFQDKEHTRTLALQQLSESVMDALSSLVGAQTAARGAHKLFHTLQHTTHNKQLFYELFELVLIEVFPELKRYYN
Summary
Uniprot
A0A2W1BQ22
A0A2H1VBC5
H9IUG0
A0A194PJD3
A0A212FHX5
A0A194RSR5
+ More
A0A1Y1M2I2 A0A1W4X6P7 A0A1W4XHD4 F4WL67 A0A3L8D8I3 E2BWJ5 A0A195CIA6 A0A151WRB4 A0A195EK76 A0A2A3EHZ7 A0A158NZJ3 E2AR00 A0A195BPA3 A0A0J7KWR0 A0A195FCV2 A0A154PAF5 A0A088A8Q0 A0A026VT11 A0A139WJS4 D2A205 K7ITU5 A0A1B0DM77 A0A0L7RF05 A0A1S4FII0 Q16ZY4 U4UL61 A0A0K8U8S4 A0A0A1XPH5 W8BK78 B0WZ22 A0A1B0CG91 A0A1W4VIM8 B4Q0Q5 A0A067RK77 B4IL52 B4R688 Q9W3N0 A0A1Q3FI08 Q8T4E8 A0A182GXA4 Q29I26 A0A0L0C822 B3NTU9 A0A1I8NGR9 B4NCV8 A0A1I8PMK6 B4JML8 B4L5D0 A0A0A1XHB2 A0A3B0K6W4 B4MAC5 A0A0M4ERA0 A0A1B0F9K0 A0A2J7RFM7 T1PHI8 B4H4J4 E0V953 A0A1Y9HDJ9 A0A310SEI0 A0A182LVF3 A0A182T6L9 A0A182VT22 A0A182J9Z4 A0A1A9VDL8 A0A182PL28 A0A182URB2 Q7Q4R5 A0A182XNK1 A0A182QU64 A0A182JXB7 A0A1J1HHP3 A0A1A9WDB2 A0A182NAC8 A0A034VJM8 A0A210QFZ3 T1INF6 V4AK36 A0A131YWS2 L7M0E1 A0A1E1XK12 A0A131XHD2 A0A1E1XFY0 A0A0P6ESV4 A0A0P5BYV2 A0A0P5DJS6 A0A0P5FLC6 A0A224Z4Z3 A0A0P5EFZ0 A0A147BGH5
A0A1Y1M2I2 A0A1W4X6P7 A0A1W4XHD4 F4WL67 A0A3L8D8I3 E2BWJ5 A0A195CIA6 A0A151WRB4 A0A195EK76 A0A2A3EHZ7 A0A158NZJ3 E2AR00 A0A195BPA3 A0A0J7KWR0 A0A195FCV2 A0A154PAF5 A0A088A8Q0 A0A026VT11 A0A139WJS4 D2A205 K7ITU5 A0A1B0DM77 A0A0L7RF05 A0A1S4FII0 Q16ZY4 U4UL61 A0A0K8U8S4 A0A0A1XPH5 W8BK78 B0WZ22 A0A1B0CG91 A0A1W4VIM8 B4Q0Q5 A0A067RK77 B4IL52 B4R688 Q9W3N0 A0A1Q3FI08 Q8T4E8 A0A182GXA4 Q29I26 A0A0L0C822 B3NTU9 A0A1I8NGR9 B4NCV8 A0A1I8PMK6 B4JML8 B4L5D0 A0A0A1XHB2 A0A3B0K6W4 B4MAC5 A0A0M4ERA0 A0A1B0F9K0 A0A2J7RFM7 T1PHI8 B4H4J4 E0V953 A0A1Y9HDJ9 A0A310SEI0 A0A182LVF3 A0A182T6L9 A0A182VT22 A0A182J9Z4 A0A1A9VDL8 A0A182PL28 A0A182URB2 Q7Q4R5 A0A182XNK1 A0A182QU64 A0A182JXB7 A0A1J1HHP3 A0A1A9WDB2 A0A182NAC8 A0A034VJM8 A0A210QFZ3 T1INF6 V4AK36 A0A131YWS2 L7M0E1 A0A1E1XK12 A0A131XHD2 A0A1E1XFY0 A0A0P6ESV4 A0A0P5BYV2 A0A0P5DJS6 A0A0P5FLC6 A0A224Z4Z3 A0A0P5EFZ0 A0A147BGH5
Pubmed
28756777
19121390
26354079
22118469
28004739
21719571
+ More
30249741 20798317 21347285 24508170 18362917 19820115 20075255 17510324 23537049 25830018 24495485 17994087 17550304 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 26108605 25315136 20566863 12364791 25348373 28812685 23254933 26830274 25576852 29209593 28049606 28503490 28797301 29652888
30249741 20798317 21347285 24508170 18362917 19820115 20075255 17510324 23537049 25830018 24495485 17994087 17550304 24845553 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 15632085 26108605 25315136 20566863 12364791 25348373 28812685 23254933 26830274 25576852 29209593 28049606 28503490 28797301 29652888
EMBL
KZ149987
PZC75665.1
ODYU01001638
SOQ38143.1
BABH01001081
KQ459606
+ More
KPI91195.1 AGBW02008449 OWR53340.1 KQ459700 KPJ20390.1 GEZM01042356 JAV79913.1 GL888207 EGI65030.1 QOIP01000012 RLU16238.1 GL451131 EFN79951.1 KQ977720 KYN00466.1 KQ982807 KYQ50390.1 KQ978747 KYN28653.1 KZ288254 PBC30892.1 ADTU01004645 GL441846 EFN64146.1 KQ976433 KYM87284.1 LBMM01002332 KMQ94967.1 KQ981685 KYN38027.1 KQ434844 KZC08170.1 KK108267 EZA46800.1 KQ971338 KYB28172.1 EFA01504.2 AJVK01006884 AJVK01006885 KQ414609 KOC69316.1 CH477480 EAT40240.1 KB632374 ERL93827.1 GDHF01029252 JAI23062.1 GBXI01001475 JAD12817.1 GAMC01016569 GAMC01016566 JAB89989.1 DS232203 EDS37336.1 AJWK01010859 CM000162 EDX02326.1 KK852422 KDR24261.1 CH480865 EDW53669.1 CM000366 EDX17365.1 AE014298 BT150246 AAF46292.2 AGV77148.1 AHN59464.1 GFDL01007867 JAV27178.1 AY089224 AAL89962.1 JXUM01020077 JXUM01020078 JXUM01020079 KQ560563 KXJ81886.1 CH379064 EAL31580.2 JRES01000777 KNC28406.1 CH954180 EDV47512.2 CH964239 EDW82667.1 CH916371 EDV91961.1 CH933811 EDW06389.1 GBXI01004394 JAD09898.1 OUUW01000042 SPP89957.1 CH940655 EDW66184.1 CP012528 ALC48615.1 CCAG010020064 NEVH01004410 PNF39620.1 KA648287 AFP62916.1 CH479209 EDW32745.1 DS234988 EEB09909.1 KQ767320 OAD53406.1 AXCM01002513 AAAB01008963 EAA12049.5 AXCN02001441 CVRI01000004 CRK87551.1 GAKP01015441 JAC43511.1 NEDP02003825 OWF47677.1 JH431165 KB201890 ESO93906.1 GEDV01005555 JAP83002.1 GACK01008490 JAA56544.1 GFAA01003881 JAT99553.1 GEFH01002122 JAP66459.1 GFAC01000998 JAT98190.1 GDIQ01071838 JAN22899.1 GDIP01178084 GDIP01178083 GDIP01178082 JAJ45320.1 GDIP01201515 GDIP01164724 GDIP01158578 GDIP01141220 GDIP01092225 JAJ64824.1 GDIQ01255322 GDIQ01229030 GDIQ01128316 JAJ96402.1 GFPF01011993 MAA23139.1 GDIP01232470 GDIP01231169 GDIP01231168 GDIP01229017 GDIP01202924 GDIP01199062 GDIP01148922 GDIP01148921 GDIP01148920 GDIP01148919 GDIP01147580 GDIP01147579 GDIP01147578 GDIP01147577 GDIP01141223 GDIP01141222 GDIP01141221 GDIP01137977 GDIP01137976 GDIP01137379 GDIP01133288 GDIP01133287 GDIP01132132 GDIP01120826 JAJ74483.1 GEGO01005570 JAR89834.1
KPI91195.1 AGBW02008449 OWR53340.1 KQ459700 KPJ20390.1 GEZM01042356 JAV79913.1 GL888207 EGI65030.1 QOIP01000012 RLU16238.1 GL451131 EFN79951.1 KQ977720 KYN00466.1 KQ982807 KYQ50390.1 KQ978747 KYN28653.1 KZ288254 PBC30892.1 ADTU01004645 GL441846 EFN64146.1 KQ976433 KYM87284.1 LBMM01002332 KMQ94967.1 KQ981685 KYN38027.1 KQ434844 KZC08170.1 KK108267 EZA46800.1 KQ971338 KYB28172.1 EFA01504.2 AJVK01006884 AJVK01006885 KQ414609 KOC69316.1 CH477480 EAT40240.1 KB632374 ERL93827.1 GDHF01029252 JAI23062.1 GBXI01001475 JAD12817.1 GAMC01016569 GAMC01016566 JAB89989.1 DS232203 EDS37336.1 AJWK01010859 CM000162 EDX02326.1 KK852422 KDR24261.1 CH480865 EDW53669.1 CM000366 EDX17365.1 AE014298 BT150246 AAF46292.2 AGV77148.1 AHN59464.1 GFDL01007867 JAV27178.1 AY089224 AAL89962.1 JXUM01020077 JXUM01020078 JXUM01020079 KQ560563 KXJ81886.1 CH379064 EAL31580.2 JRES01000777 KNC28406.1 CH954180 EDV47512.2 CH964239 EDW82667.1 CH916371 EDV91961.1 CH933811 EDW06389.1 GBXI01004394 JAD09898.1 OUUW01000042 SPP89957.1 CH940655 EDW66184.1 CP012528 ALC48615.1 CCAG010020064 NEVH01004410 PNF39620.1 KA648287 AFP62916.1 CH479209 EDW32745.1 DS234988 EEB09909.1 KQ767320 OAD53406.1 AXCM01002513 AAAB01008963 EAA12049.5 AXCN02001441 CVRI01000004 CRK87551.1 GAKP01015441 JAC43511.1 NEDP02003825 OWF47677.1 JH431165 KB201890 ESO93906.1 GEDV01005555 JAP83002.1 GACK01008490 JAA56544.1 GFAA01003881 JAT99553.1 GEFH01002122 JAP66459.1 GFAC01000998 JAT98190.1 GDIQ01071838 JAN22899.1 GDIP01178084 GDIP01178083 GDIP01178082 JAJ45320.1 GDIP01201515 GDIP01164724 GDIP01158578 GDIP01141220 GDIP01092225 JAJ64824.1 GDIQ01255322 GDIQ01229030 GDIQ01128316 JAJ96402.1 GFPF01011993 MAA23139.1 GDIP01232470 GDIP01231169 GDIP01231168 GDIP01229017 GDIP01202924 GDIP01199062 GDIP01148922 GDIP01148921 GDIP01148920 GDIP01148919 GDIP01147580 GDIP01147579 GDIP01147578 GDIP01147577 GDIP01141223 GDIP01141222 GDIP01141221 GDIP01137977 GDIP01137976 GDIP01137379 GDIP01133288 GDIP01133287 GDIP01132132 GDIP01120826 JAJ74483.1 GEGO01005570 JAR89834.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000192223
UP000007755
+ More
UP000279307 UP000008237 UP000078542 UP000075809 UP000078492 UP000242457 UP000005205 UP000000311 UP000078540 UP000036403 UP000078541 UP000076502 UP000005203 UP000053097 UP000007266 UP000002358 UP000092462 UP000053825 UP000008820 UP000030742 UP000002320 UP000092461 UP000192221 UP000002282 UP000027135 UP000001292 UP000000304 UP000000803 UP000069940 UP000249989 UP000001819 UP000037069 UP000008711 UP000095301 UP000007798 UP000095300 UP000001070 UP000009192 UP000268350 UP000008792 UP000092553 UP000092444 UP000235965 UP000008744 UP000009046 UP000075900 UP000075883 UP000075901 UP000075920 UP000075880 UP000078200 UP000075885 UP000075903 UP000007062 UP000076407 UP000075886 UP000075881 UP000183832 UP000091820 UP000075884 UP000242188 UP000030746
UP000279307 UP000008237 UP000078542 UP000075809 UP000078492 UP000242457 UP000005205 UP000000311 UP000078540 UP000036403 UP000078541 UP000076502 UP000005203 UP000053097 UP000007266 UP000002358 UP000092462 UP000053825 UP000008820 UP000030742 UP000002320 UP000092461 UP000192221 UP000002282 UP000027135 UP000001292 UP000000304 UP000000803 UP000069940 UP000249989 UP000001819 UP000037069 UP000008711 UP000095301 UP000007798 UP000095300 UP000001070 UP000009192 UP000268350 UP000008792 UP000092553 UP000092444 UP000235965 UP000008744 UP000009046 UP000075900 UP000075883 UP000075901 UP000075920 UP000075880 UP000078200 UP000075885 UP000075903 UP000007062 UP000076407 UP000075886 UP000075881 UP000183832 UP000091820 UP000075884 UP000242188 UP000030746
Pfam
Interpro
IPR003114
Phox_assoc
+ More
IPR037899 SNX25_PX
IPR013937 Sorting_nexin_C
IPR016137 RGS
IPR001683 Phox
IPR036871 PX_dom_sf
IPR036305 RGS_sf
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR002129 PyrdxlP-dep_de-COase
IPR019172 Osteopetrosis-assoc_TM_1
IPR013087 Znf_C2H2_type
IPR009360 Isy1
IPR034905 SNX25
IPR037899 SNX25_PX
IPR013937 Sorting_nexin_C
IPR016137 RGS
IPR001683 Phox
IPR036871 PX_dom_sf
IPR036305 RGS_sf
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR002129 PyrdxlP-dep_de-COase
IPR019172 Osteopetrosis-assoc_TM_1
IPR013087 Znf_C2H2_type
IPR009360 Isy1
IPR034905 SNX25
Gene 3D
CDD
ProteinModelPortal
A0A2W1BQ22
A0A2H1VBC5
H9IUG0
A0A194PJD3
A0A212FHX5
A0A194RSR5
+ More
A0A1Y1M2I2 A0A1W4X6P7 A0A1W4XHD4 F4WL67 A0A3L8D8I3 E2BWJ5 A0A195CIA6 A0A151WRB4 A0A195EK76 A0A2A3EHZ7 A0A158NZJ3 E2AR00 A0A195BPA3 A0A0J7KWR0 A0A195FCV2 A0A154PAF5 A0A088A8Q0 A0A026VT11 A0A139WJS4 D2A205 K7ITU5 A0A1B0DM77 A0A0L7RF05 A0A1S4FII0 Q16ZY4 U4UL61 A0A0K8U8S4 A0A0A1XPH5 W8BK78 B0WZ22 A0A1B0CG91 A0A1W4VIM8 B4Q0Q5 A0A067RK77 B4IL52 B4R688 Q9W3N0 A0A1Q3FI08 Q8T4E8 A0A182GXA4 Q29I26 A0A0L0C822 B3NTU9 A0A1I8NGR9 B4NCV8 A0A1I8PMK6 B4JML8 B4L5D0 A0A0A1XHB2 A0A3B0K6W4 B4MAC5 A0A0M4ERA0 A0A1B0F9K0 A0A2J7RFM7 T1PHI8 B4H4J4 E0V953 A0A1Y9HDJ9 A0A310SEI0 A0A182LVF3 A0A182T6L9 A0A182VT22 A0A182J9Z4 A0A1A9VDL8 A0A182PL28 A0A182URB2 Q7Q4R5 A0A182XNK1 A0A182QU64 A0A182JXB7 A0A1J1HHP3 A0A1A9WDB2 A0A182NAC8 A0A034VJM8 A0A210QFZ3 T1INF6 V4AK36 A0A131YWS2 L7M0E1 A0A1E1XK12 A0A131XHD2 A0A1E1XFY0 A0A0P6ESV4 A0A0P5BYV2 A0A0P5DJS6 A0A0P5FLC6 A0A224Z4Z3 A0A0P5EFZ0 A0A147BGH5
A0A1Y1M2I2 A0A1W4X6P7 A0A1W4XHD4 F4WL67 A0A3L8D8I3 E2BWJ5 A0A195CIA6 A0A151WRB4 A0A195EK76 A0A2A3EHZ7 A0A158NZJ3 E2AR00 A0A195BPA3 A0A0J7KWR0 A0A195FCV2 A0A154PAF5 A0A088A8Q0 A0A026VT11 A0A139WJS4 D2A205 K7ITU5 A0A1B0DM77 A0A0L7RF05 A0A1S4FII0 Q16ZY4 U4UL61 A0A0K8U8S4 A0A0A1XPH5 W8BK78 B0WZ22 A0A1B0CG91 A0A1W4VIM8 B4Q0Q5 A0A067RK77 B4IL52 B4R688 Q9W3N0 A0A1Q3FI08 Q8T4E8 A0A182GXA4 Q29I26 A0A0L0C822 B3NTU9 A0A1I8NGR9 B4NCV8 A0A1I8PMK6 B4JML8 B4L5D0 A0A0A1XHB2 A0A3B0K6W4 B4MAC5 A0A0M4ERA0 A0A1B0F9K0 A0A2J7RFM7 T1PHI8 B4H4J4 E0V953 A0A1Y9HDJ9 A0A310SEI0 A0A182LVF3 A0A182T6L9 A0A182VT22 A0A182J9Z4 A0A1A9VDL8 A0A182PL28 A0A182URB2 Q7Q4R5 A0A182XNK1 A0A182QU64 A0A182JXB7 A0A1J1HHP3 A0A1A9WDB2 A0A182NAC8 A0A034VJM8 A0A210QFZ3 T1INF6 V4AK36 A0A131YWS2 L7M0E1 A0A1E1XK12 A0A131XHD2 A0A1E1XFY0 A0A0P6ESV4 A0A0P5BYV2 A0A0P5DJS6 A0A0P5FLC6 A0A224Z4Z3 A0A0P5EFZ0 A0A147BGH5
PDB
5WOE
E-value=1.50048e-16,
Score=215
Ontologies
GO
PANTHER
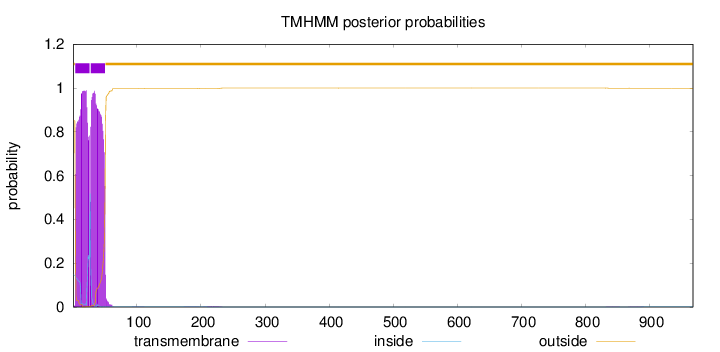
Topology
Length:
968
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.5685700000002
Exp number, first 60 AAs:
41.47397
Total prob of N-in:
0.14887
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 27
TMhelix
28 - 50
outside
51 - 968
Population Genetic Test Statistics
Pi
251.901615
Theta
158.851919
Tajima's D
1.313198
CLR
0.199801
CSRT
0.741212939353032
Interpretation
Uncertain
