Gene
KWMTBOMO07749
Pre Gene Modal
BGIBMGA001257
Annotation
Pyridine_nucleotide-disulfide_oxidoreductase_domain-containing_protein_1_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 3.12
Sequence
CDS
ATGTCGGAGATTAATACTACATATTTAATAGTTGGCGGTGGTATTGCTGGAGTTACTTGCGTAGAAACTCTTGCGATCTTGCATCCTGAAAAAACCTTGGTACTAGTAACAGCATCAACACTTGTGAAAAATGTTAGTAACGTTACCTTTTTTGCTAAGACTATAGTGAACTTTGACGTGAATGAAACAGAGGCCACCTCCTTGCAAAAGATCCATCCGAATTTAAAAATTATATATGATTCACTAAAACATCTTGATACTGAAAAGCAGAAAGCTTTGACAGACAAAGGTGTTCATATACAATATGACGTAATTTGTATTTGTACTGGAGGGATACCAAGACTAATTTCAGATTCCAAAAATAGCAAAAGAGTACTGGGCATTAGAGATACAGAATCTGTGAAAGACTTTCAACAAAAATTGAAGACAGGCCGTAGAATGGTTATAGTTGGAAATGGAGGTGCTGCTGAGTTTTTTCAAGAAACCTTTAAAAATGAGAATGAAAGAAAAAATCAAAATACAGTTTTAAGGAGGCATATTTACTCTGAAGAAGACAGTTTGGTATCTTTAAATAAAAATTTAAAATCTGCTGCACTTGGTCCAGATTGGTATAGGAAACTTGAAAACATTAAGTCTGACAGAGGGGTCCAAGAACTCGAAATAGTATATAAAGCTGAAGTAGAATCAGTATTAGAGGATAATAAGAATGAATATCCATTACGTGTACAACTAACAAATTCTCAGCTTGTTGAGTGTGATTTTTTAATATCTGCAACTGGGGTGGAACCATCTGTCAACTTTACATGGGATAAAGTTCCAGAGAAAGGTCAGGATGGAGGATTGGCAGTCAATGAATTTCAAGAGACTAGTATAAAAAATGTATTTGCTGCAGGCGATGTTGCATCAGCTGCATGGGAACATGCCCCACACTGGTTTCAATTACGTCTCTGGACTCAAGCCAGACAAATGGCTGGAATGGCTGCTAAAGCAATGCATGGTAGAATAACAAATCAAGAAGTTTTACAAGATTTCTGTTTTGAACTGTTTACTCATTGCACAAAATTGTTTGGTTATAGAGTCATTTTATTAGGAAAATACAATGGACAAGGCCTAGGCACTGATTATGAAATTCTTCTTCGAACTACTCCTAATGTTGAATACATTAAGTTTGTATTAAAAGAAGGAAAGCTACAAGGAGCAATTTTAATAGGCGAAACTGATCTTGAAGAAATGTGTGAGAACCTGATTCTAGACCAAATTGATTTATCACCATTTGGTGATGATATCTTGAATCCAGATATAGACATTGATGATTATTTTGATTAA
Protein
MSEINTTYLIVGGGIAGVTCVETLAILHPEKTLVLVTASTLVKNVSNVTFFAKTIVNFDVNETEATSLQKIHPNLKIIYDSLKHLDTEKQKALTDKGVHIQYDVICICTGGIPRLISDSKNSKRVLGIRDTESVKDFQQKLKTGRRMVIVGNGGAAEFFQETFKNENERKNQNTVLRRHIYSEEDSLVSLNKNLKSAALGPDWYRKLENIKSDRGVQELEIVYKAEVESVLEDNKNEYPLRVQLTNSQLVECDFLISATGVEPSVNFTWDKVPEKGQDGGLAVNEFQETSIKNVFAAGDVASAAWEHAPHWFQLRLWTQARQMAGMAAKAMHGRITNQEVLQDFCFELFTHCTKLFGYRVILLGKYNGQGLGTDYEILLRTTPNVEYIKFVLKEGKLQGAILIGETDLEEMCENLILDQIDLSPFGDDILNPDIDIDDYFD
Summary
Uniprot
H9IVH7
A0A2W1BSP7
A0A0L7LIP5
A0A2A4JWN4
A0A2H1VBD5
A0A194RW24
+ More
A0A194PES1 A0A212FHY2 S4P998 A0A182GSK6 A0A023ESP3 W5JPU6 A0A2M4AMI0 A0A182F618 U5EV03 A0A2M4AMS4 A0A182QHR5 W8BQ04 A0A1Q3FHV2 A0A0K8TMX8 Q17JG6 A0A034V168 A0A2M4BL21 A0A0T6AW28 A0A182P4V5 A0A2M4BKW6 A0A182L1Q2 A0A2M4BMI7 A0A182VJ23 A0A067QI01 A0A182UCL6 Q7Q3T9 B0W7M8 A0A2P8YGQ5 A0A0K8W0H9 A0A088AAI3 A0A195CD51 A0A1B6GN58 T1H193 A0A0M9A619 A0A336K4F7 A0A182WUA1 A0A2J7QNL1 A0A2A3E1V3 V9IM63 A0A195FQY3 E2A5Q5 A0A1B6IQ17 A0A1Y1M187 A0A1B6J7I9 A0A182NTR7 D6WL78 K7IRH1 A0A0A9XRI0 A0A182MAK0 A0A146LWL0 A0A1L8DZV9 A0A182W7A6 A0A0J7KNF0 A0A182SYU7 A0A195DMX6 A0A154PCB9 A0A151X7Q3 E9IH74 E2B897 A0A0K8SBP7 F4X1F4 A0A182RC79 A0A182JR63 A0A195BKG2 A0A2R7VRW2 A0A182JIJ6 A0A026WQ93 A0A2R2MTT3 A0A0L7RI72 A0A1B6MII5 A0A1W4WFL3 A0A158NA84 A0A1I8PN84 A0A0A1XSS7 A0A224XNA7 A0A084VNE1 E0VJS8 Q29KL2 R4WPW3 A0A1L8DZE1 R4FNT7 A0A0P4VS51 A0A1B0G2F7 A0A210QJQ7 A0A1B0C695 A0A0M5IYS1 A0A1B0AKN2 A0A1B0AH67 T1PCT2 B3RS58 A0A1B0DM11 A0A2M4ANW9 A0A1A9YRT4 A0A0K8SBP4
A0A194PES1 A0A212FHY2 S4P998 A0A182GSK6 A0A023ESP3 W5JPU6 A0A2M4AMI0 A0A182F618 U5EV03 A0A2M4AMS4 A0A182QHR5 W8BQ04 A0A1Q3FHV2 A0A0K8TMX8 Q17JG6 A0A034V168 A0A2M4BL21 A0A0T6AW28 A0A182P4V5 A0A2M4BKW6 A0A182L1Q2 A0A2M4BMI7 A0A182VJ23 A0A067QI01 A0A182UCL6 Q7Q3T9 B0W7M8 A0A2P8YGQ5 A0A0K8W0H9 A0A088AAI3 A0A195CD51 A0A1B6GN58 T1H193 A0A0M9A619 A0A336K4F7 A0A182WUA1 A0A2J7QNL1 A0A2A3E1V3 V9IM63 A0A195FQY3 E2A5Q5 A0A1B6IQ17 A0A1Y1M187 A0A1B6J7I9 A0A182NTR7 D6WL78 K7IRH1 A0A0A9XRI0 A0A182MAK0 A0A146LWL0 A0A1L8DZV9 A0A182W7A6 A0A0J7KNF0 A0A182SYU7 A0A195DMX6 A0A154PCB9 A0A151X7Q3 E9IH74 E2B897 A0A0K8SBP7 F4X1F4 A0A182RC79 A0A182JR63 A0A195BKG2 A0A2R7VRW2 A0A182JIJ6 A0A026WQ93 A0A2R2MTT3 A0A0L7RI72 A0A1B6MII5 A0A1W4WFL3 A0A158NA84 A0A1I8PN84 A0A0A1XSS7 A0A224XNA7 A0A084VNE1 E0VJS8 Q29KL2 R4WPW3 A0A1L8DZE1 R4FNT7 A0A0P4VS51 A0A1B0G2F7 A0A210QJQ7 A0A1B0C695 A0A0M5IYS1 A0A1B0AKN2 A0A1B0AH67 T1PCT2 B3RS58 A0A1B0DM11 A0A2M4ANW9 A0A1A9YRT4 A0A0K8SBP4
Pubmed
19121390
28756777
26227816
26354079
22118469
23622113
+ More
26483478 24945155 20920257 23761445 24495485 26369729 17510324 25348373 20966253 24845553 12364791 14747013 17210077 29403074 20798317 28004739 18362917 19820115 20075255 25401762 26823975 21282665 21719571 24508170 30249741 21347285 25830018 24438588 20566863 15632085 17994087 23691247 27129103 28812685 18719581
26483478 24945155 20920257 23761445 24495485 26369729 17510324 25348373 20966253 24845553 12364791 14747013 17210077 29403074 20798317 28004739 18362917 19820115 20075255 25401762 26823975 21282665 21719571 24508170 30249741 21347285 25830018 24438588 20566863 15632085 17994087 23691247 27129103 28812685 18719581
EMBL
BABH01001082
KZ149987
PZC75666.1
JTDY01000982
KOB75209.1
NWSH01000440
+ More
PCG76411.1 ODYU01001638 SOQ38145.1 KQ459700 KPJ20391.1 KQ459606 KPI91194.1 AGBW02008449 OWR53339.1 GAIX01003834 JAA88726.1 JXUM01016999 KQ560472 KXJ82230.1 GAPW01001288 JAC12310.1 ADMH02000877 ETN64789.1 GGFK01008611 MBW41932.1 GANO01001230 JAB58641.1 GGFK01008760 MBW42081.1 AXCN02000801 GAMC01007658 GAMC01007657 GAMC01007655 GAMC01007653 JAB98898.1 GFDL01007894 JAV27151.1 GDAI01002090 JAI15513.1 CH477232 EAT46874.1 GAKP01023418 JAC35540.1 GGFJ01004553 MBW53694.1 LJIG01022669 KRT79338.1 GGFJ01004554 MBW53695.1 GGFJ01005032 MBW54173.1 KK853366 KDR08097.1 AAAB01008964 EAA12697.3 DS231854 EDS38063.1 PYGN01000608 PSN43440.1 GDHF01033092 GDHF01007730 GDHF01006091 JAI19222.1 JAI44584.1 JAI46223.1 KQ978009 KYM98141.1 GECZ01006006 JAS63763.1 CAQQ02162574 CAQQ02162575 KQ435727 KOX77967.1 UFQS01000114 UFQT01000114 SSW99854.1 SSX20234.1 NEVH01013194 PNF30168.1 KZ288444 PBC25680.1 JR053022 AEY61842.1 KQ981305 KYN42995.1 GL437023 EFN71252.1 GECU01018729 JAS88977.1 GEZM01045892 JAV77636.1 GECU01012560 JAS95146.1 KQ971343 EFA04075.1 GBHO01021040 GBHO01021039 JAG22564.1 JAG22565.1 AXCM01001558 GDHC01006606 JAQ12023.1 GFDF01002252 JAV11832.1 LBMM01005165 KMQ91769.1 KQ980734 KYN13854.1 KQ434870 KZC09569.1 KQ982446 KYQ56340.1 GL763190 EFZ20079.1 GL446286 EFN88099.1 GBRD01015095 JAG50731.1 GL888529 EGI59682.1 KQ976455 KYM85172.1 KK854016 PTY09410.1 KK107131 QOIP01000012 EZA58180.1 RLU15619.1 KQ414585 KOC70524.1 GEBQ01021348 GEBQ01004308 JAT18629.1 JAT35669.1 ADTU01010075 GBXI01014518 GBXI01000211 JAC99773.1 JAD14081.1 GFTR01006506 JAW09920.1 ATLV01014737 KE524984 KFB39485.1 AAZO01002894 DS235226 EEB13634.1 CH379061 EAL33162.1 AK417716 BAN20931.1 GFDF01002253 JAV11831.1 ACPB03001438 GAHY01000443 JAA77067.1 GDKW01001327 JAI55268.1 CCAG010014651 NEDP02003316 OWF48998.1 JXJN01026508 CP012523 ALC39998.1 JXJN01026397 KA645940 AFP60569.1 DS985243 EDV26459.1 AJVK01006815 GGFK01009158 MBW42479.1 GBRD01015096 GBRD01015094 JAG50732.1
PCG76411.1 ODYU01001638 SOQ38145.1 KQ459700 KPJ20391.1 KQ459606 KPI91194.1 AGBW02008449 OWR53339.1 GAIX01003834 JAA88726.1 JXUM01016999 KQ560472 KXJ82230.1 GAPW01001288 JAC12310.1 ADMH02000877 ETN64789.1 GGFK01008611 MBW41932.1 GANO01001230 JAB58641.1 GGFK01008760 MBW42081.1 AXCN02000801 GAMC01007658 GAMC01007657 GAMC01007655 GAMC01007653 JAB98898.1 GFDL01007894 JAV27151.1 GDAI01002090 JAI15513.1 CH477232 EAT46874.1 GAKP01023418 JAC35540.1 GGFJ01004553 MBW53694.1 LJIG01022669 KRT79338.1 GGFJ01004554 MBW53695.1 GGFJ01005032 MBW54173.1 KK853366 KDR08097.1 AAAB01008964 EAA12697.3 DS231854 EDS38063.1 PYGN01000608 PSN43440.1 GDHF01033092 GDHF01007730 GDHF01006091 JAI19222.1 JAI44584.1 JAI46223.1 KQ978009 KYM98141.1 GECZ01006006 JAS63763.1 CAQQ02162574 CAQQ02162575 KQ435727 KOX77967.1 UFQS01000114 UFQT01000114 SSW99854.1 SSX20234.1 NEVH01013194 PNF30168.1 KZ288444 PBC25680.1 JR053022 AEY61842.1 KQ981305 KYN42995.1 GL437023 EFN71252.1 GECU01018729 JAS88977.1 GEZM01045892 JAV77636.1 GECU01012560 JAS95146.1 KQ971343 EFA04075.1 GBHO01021040 GBHO01021039 JAG22564.1 JAG22565.1 AXCM01001558 GDHC01006606 JAQ12023.1 GFDF01002252 JAV11832.1 LBMM01005165 KMQ91769.1 KQ980734 KYN13854.1 KQ434870 KZC09569.1 KQ982446 KYQ56340.1 GL763190 EFZ20079.1 GL446286 EFN88099.1 GBRD01015095 JAG50731.1 GL888529 EGI59682.1 KQ976455 KYM85172.1 KK854016 PTY09410.1 KK107131 QOIP01000012 EZA58180.1 RLU15619.1 KQ414585 KOC70524.1 GEBQ01021348 GEBQ01004308 JAT18629.1 JAT35669.1 ADTU01010075 GBXI01014518 GBXI01000211 JAC99773.1 JAD14081.1 GFTR01006506 JAW09920.1 ATLV01014737 KE524984 KFB39485.1 AAZO01002894 DS235226 EEB13634.1 CH379061 EAL33162.1 AK417716 BAN20931.1 GFDF01002253 JAV11831.1 ACPB03001438 GAHY01000443 JAA77067.1 GDKW01001327 JAI55268.1 CCAG010014651 NEDP02003316 OWF48998.1 JXJN01026508 CP012523 ALC39998.1 JXJN01026397 KA645940 AFP60569.1 DS985243 EDV26459.1 AJVK01006815 GGFK01009158 MBW42479.1 GBRD01015096 GBRD01015094 JAG50732.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000069940 UP000249989 UP000000673 UP000069272 UP000075886 UP000008820 UP000075885 UP000075882 UP000075903 UP000027135 UP000075902 UP000007062 UP000002320 UP000245037 UP000005203 UP000078542 UP000015102 UP000053105 UP000076407 UP000235965 UP000242457 UP000078541 UP000000311 UP000075884 UP000007266 UP000002358 UP000075883 UP000075920 UP000036403 UP000075901 UP000078492 UP000076502 UP000075809 UP000008237 UP000007755 UP000075900 UP000075881 UP000078540 UP000075880 UP000053097 UP000279307 UP000085678 UP000053825 UP000192223 UP000005205 UP000095300 UP000030765 UP000009046 UP000001819 UP000015103 UP000092444 UP000242188 UP000092460 UP000092553 UP000092445 UP000009022 UP000092462 UP000092443
UP000069940 UP000249989 UP000000673 UP000069272 UP000075886 UP000008820 UP000075885 UP000075882 UP000075903 UP000027135 UP000075902 UP000007062 UP000002320 UP000245037 UP000005203 UP000078542 UP000015102 UP000053105 UP000076407 UP000235965 UP000242457 UP000078541 UP000000311 UP000075884 UP000007266 UP000002358 UP000075883 UP000075920 UP000036403 UP000075901 UP000078492 UP000076502 UP000075809 UP000008237 UP000007755 UP000075900 UP000075881 UP000078540 UP000075880 UP000053097 UP000279307 UP000085678 UP000053825 UP000192223 UP000005205 UP000095300 UP000030765 UP000009046 UP000001819 UP000015103 UP000092444 UP000242188 UP000092460 UP000092553 UP000092445 UP000009022 UP000092462 UP000092443
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9IVH7
A0A2W1BSP7
A0A0L7LIP5
A0A2A4JWN4
A0A2H1VBD5
A0A194RW24
+ More
A0A194PES1 A0A212FHY2 S4P998 A0A182GSK6 A0A023ESP3 W5JPU6 A0A2M4AMI0 A0A182F618 U5EV03 A0A2M4AMS4 A0A182QHR5 W8BQ04 A0A1Q3FHV2 A0A0K8TMX8 Q17JG6 A0A034V168 A0A2M4BL21 A0A0T6AW28 A0A182P4V5 A0A2M4BKW6 A0A182L1Q2 A0A2M4BMI7 A0A182VJ23 A0A067QI01 A0A182UCL6 Q7Q3T9 B0W7M8 A0A2P8YGQ5 A0A0K8W0H9 A0A088AAI3 A0A195CD51 A0A1B6GN58 T1H193 A0A0M9A619 A0A336K4F7 A0A182WUA1 A0A2J7QNL1 A0A2A3E1V3 V9IM63 A0A195FQY3 E2A5Q5 A0A1B6IQ17 A0A1Y1M187 A0A1B6J7I9 A0A182NTR7 D6WL78 K7IRH1 A0A0A9XRI0 A0A182MAK0 A0A146LWL0 A0A1L8DZV9 A0A182W7A6 A0A0J7KNF0 A0A182SYU7 A0A195DMX6 A0A154PCB9 A0A151X7Q3 E9IH74 E2B897 A0A0K8SBP7 F4X1F4 A0A182RC79 A0A182JR63 A0A195BKG2 A0A2R7VRW2 A0A182JIJ6 A0A026WQ93 A0A2R2MTT3 A0A0L7RI72 A0A1B6MII5 A0A1W4WFL3 A0A158NA84 A0A1I8PN84 A0A0A1XSS7 A0A224XNA7 A0A084VNE1 E0VJS8 Q29KL2 R4WPW3 A0A1L8DZE1 R4FNT7 A0A0P4VS51 A0A1B0G2F7 A0A210QJQ7 A0A1B0C695 A0A0M5IYS1 A0A1B0AKN2 A0A1B0AH67 T1PCT2 B3RS58 A0A1B0DM11 A0A2M4ANW9 A0A1A9YRT4 A0A0K8SBP4
A0A194PES1 A0A212FHY2 S4P998 A0A182GSK6 A0A023ESP3 W5JPU6 A0A2M4AMI0 A0A182F618 U5EV03 A0A2M4AMS4 A0A182QHR5 W8BQ04 A0A1Q3FHV2 A0A0K8TMX8 Q17JG6 A0A034V168 A0A2M4BL21 A0A0T6AW28 A0A182P4V5 A0A2M4BKW6 A0A182L1Q2 A0A2M4BMI7 A0A182VJ23 A0A067QI01 A0A182UCL6 Q7Q3T9 B0W7M8 A0A2P8YGQ5 A0A0K8W0H9 A0A088AAI3 A0A195CD51 A0A1B6GN58 T1H193 A0A0M9A619 A0A336K4F7 A0A182WUA1 A0A2J7QNL1 A0A2A3E1V3 V9IM63 A0A195FQY3 E2A5Q5 A0A1B6IQ17 A0A1Y1M187 A0A1B6J7I9 A0A182NTR7 D6WL78 K7IRH1 A0A0A9XRI0 A0A182MAK0 A0A146LWL0 A0A1L8DZV9 A0A182W7A6 A0A0J7KNF0 A0A182SYU7 A0A195DMX6 A0A154PCB9 A0A151X7Q3 E9IH74 E2B897 A0A0K8SBP7 F4X1F4 A0A182RC79 A0A182JR63 A0A195BKG2 A0A2R7VRW2 A0A182JIJ6 A0A026WQ93 A0A2R2MTT3 A0A0L7RI72 A0A1B6MII5 A0A1W4WFL3 A0A158NA84 A0A1I8PN84 A0A0A1XSS7 A0A224XNA7 A0A084VNE1 E0VJS8 Q29KL2 R4WPW3 A0A1L8DZE1 R4FNT7 A0A0P4VS51 A0A1B0G2F7 A0A210QJQ7 A0A1B0C695 A0A0M5IYS1 A0A1B0AKN2 A0A1B0AH67 T1PCT2 B3RS58 A0A1B0DM11 A0A2M4ANW9 A0A1A9YRT4 A0A0K8SBP4
PDB
5JCN
E-value=1.44026e-06,
Score=125
Ontologies
GO
Topology
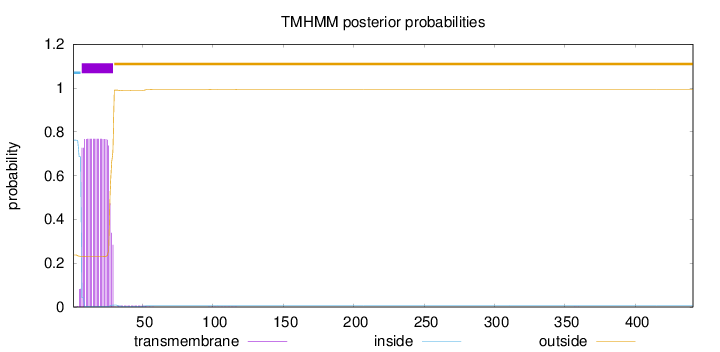
Length:
441
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.66528
Exp number, first 60 AAs:
16.64278
Total prob of N-in:
0.76214
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 441
Population Genetic Test Statistics
Pi
126.368773
Theta
133.193917
Tajima's D
-0.634295
CLR
169.004171
CSRT
0.209739513024349
Interpretation
Uncertain
