Pre Gene Modal
BGIBMGA001258
Annotation
PREDICTED:_synaptobrevin_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.83
Sequence
CDS
ATGCCTATATTATTCAGTATCGTGGCTAGAGGCACTGTAGTCCTCGCTAAATATGCAACTTGTCAGGGTAATTTCACTGAAGTCGCAGAGCAGATTTTATCAAAGATTCCGCCCCACGACGATAAGTTGACATATTCTCACGGAAATTATCTATTCCATTACATAGCAGAGAACAAATTGGTTTATTTCTGCATCACTGACGATAAATTCCAACGTTCGAGAGCGTTCTTATTTCTTAATGAGATAAAAAGAAGATTCACTACAGCATTTGCTGACACCGCACAGACAGCAATACCTTACGCCATGAACAGTGAATTTGGTCGGGTTCTTGCCACTGAGATGAAACATTACAGTGAATCTCGAGACTTGGACAACATTTCAAGAGTTCATGGAGAATTAGATGAATTAAAAAATATAATGGTTAAAAACATTGATAACATGGCAATGCGTGGAGAAAAGCTAGAACTGCTTGTGAACAAAGCTGATAACTTAGCCACTAGTTCAGTGTCGTATCGCGCTACTTCTCGCACACTTCAACGCTCTCTATTCTGGAAGAATATCAAAATGTATGTCATATTGGTGTCCATAGCAGCATTGGCGGTTTACTTAATAGGAGCGATGGCGTGTGGAGGCCTCGCATGGAAATCTTGCGTCGGTTAA
Protein
MPILFSIVARGTVVLAKYATCQGNFTEVAEQILSKIPPHDDKLTYSHGNYLFHYIAENKLVYFCITDDKFQRSRAFLFLNEIKRRFTTAFADTAQTAIPYAMNSEFGRVLATEMKHYSESRDLDNISRVHGELDELKNIMVKNIDNMAMRGEKLELLVNKADNLATSSVSYRATSRTLQRSLFWKNIKMYVILVSIAALAVYLIGAMACGGLAWKSCVG
Summary
Similarity
Belongs to the synaptobrevin family.
Uniprot
H9IVH8
Q157R9
A0A194PDU7
A0A212FHX6
A0A2W1BR95
A0A2A4JYF3
+ More
A0A2H1VBH0 A0A194RSE0 E2BVD4 A0A067R381 A0A1W4XJM7 A0A0C9QUM8 J3JY72 N6UIR5 A0A232FG65 A0A1Y1MUI8 A0A1B6FRE5 A0A158NBH3 F4X8P9 A0A195FDX6 E9J188 A0A195DSB8 A0A0J7KWZ1 A0A1B6KPD5 E1ZZ68 A0A1B6I2Y0 A0A2P8XEF4 E0VXM7 A0A1B6E5M8 A0A151IGU3 A0A2J7QB94 A0A1B0GKF3 A0A154NZA9 A0A1L8DVT3 A0A1B0DFU5 A0A1Q3FGT9 A0A1Q3FGV5 A0A1Q3FHS5 U5EV63 A0A182GDX2 A0A023EKR7 A0A3R7Q583 A0A1Q3FHI9 B0WWI3 A0A224XX87 A0A0V0G840 A0A069DPN0 A0A0L0CGA7 A0A336MC22 B4NWL5 B3N6V4 B4HT51 B4QHR8 A0A1W4ULZ3 Q7JYX5 A0A2A3ESM8 V9IET7 A0A088AUK2 A0A0P6IEN8 A0A1A9W6F0 A0A0P4X8A1 A0A1A9YKG9 A0A1B0BZD1 A0A0P4VZ58 A0A1J1HYD7 A0A1A9ZHY5 A0A087UNC0 A0A0P4VTT1 T1IC92 A0A1A9USD0 D3TLF4 A7SBE8 A0A2B4SX42 A0A182ME07 E9FRH1 A0A182W671 A0A1D2N7V5 B4KMG9 A0A182PV13 A0A182TGI1 A0A182S9L1 A0A3M6TZW8 A0A182XY95 A0A182VN60 A0A182X7J7 F5HJV6 A0A182I9T9 A0A1I8MIC1 C3YRU5 A0A091WH27 T1JBQ6 A0A182MZX9 A0A182QZM5 K7IZA2 A0A1S3KBD9 B4MKB4 A0A182RYU6 A0A3L8DWL2 B4LNG1 B3MIM4
A0A2H1VBH0 A0A194RSE0 E2BVD4 A0A067R381 A0A1W4XJM7 A0A0C9QUM8 J3JY72 N6UIR5 A0A232FG65 A0A1Y1MUI8 A0A1B6FRE5 A0A158NBH3 F4X8P9 A0A195FDX6 E9J188 A0A195DSB8 A0A0J7KWZ1 A0A1B6KPD5 E1ZZ68 A0A1B6I2Y0 A0A2P8XEF4 E0VXM7 A0A1B6E5M8 A0A151IGU3 A0A2J7QB94 A0A1B0GKF3 A0A154NZA9 A0A1L8DVT3 A0A1B0DFU5 A0A1Q3FGT9 A0A1Q3FGV5 A0A1Q3FHS5 U5EV63 A0A182GDX2 A0A023EKR7 A0A3R7Q583 A0A1Q3FHI9 B0WWI3 A0A224XX87 A0A0V0G840 A0A069DPN0 A0A0L0CGA7 A0A336MC22 B4NWL5 B3N6V4 B4HT51 B4QHR8 A0A1W4ULZ3 Q7JYX5 A0A2A3ESM8 V9IET7 A0A088AUK2 A0A0P6IEN8 A0A1A9W6F0 A0A0P4X8A1 A0A1A9YKG9 A0A1B0BZD1 A0A0P4VZ58 A0A1J1HYD7 A0A1A9ZHY5 A0A087UNC0 A0A0P4VTT1 T1IC92 A0A1A9USD0 D3TLF4 A7SBE8 A0A2B4SX42 A0A182ME07 E9FRH1 A0A182W671 A0A1D2N7V5 B4KMG9 A0A182PV13 A0A182TGI1 A0A182S9L1 A0A3M6TZW8 A0A182XY95 A0A182VN60 A0A182X7J7 F5HJV6 A0A182I9T9 A0A1I8MIC1 C3YRU5 A0A091WH27 T1JBQ6 A0A182MZX9 A0A182QZM5 K7IZA2 A0A1S3KBD9 B4MKB4 A0A182RYU6 A0A3L8DWL2 B4LNG1 B3MIM4
Pubmed
19121390
26354079
22118469
28756777
20798317
24845553
+ More
22516182 23537049 28648823 28004739 21347285 21719571 21282665 29403074 20566863 26483478 24945155 26334808 26108605 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 20353571 17615350 21292972 27289101 30382153 25244985 12364791 14747013 17210077 25315136 18563158 20075255 30249741 18057021
22516182 23537049 28648823 28004739 21347285 21719571 21282665 29403074 20566863 26483478 24945155 26334808 26108605 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27129103 20353571 17615350 21292972 27289101 30382153 25244985 12364791 14747013 17210077 25315136 18563158 20075255 30249741 18057021
EMBL
BABH01001082
BABH01001083
DQ539017
ABG20589.1
KQ459606
KPI91193.1
+ More
AGBW02008449 OWR53338.1 KZ149987 PZC75667.1 NWSH01000440 PCG76412.1 ODYU01001638 SOQ38146.1 KQ459700 KPJ20392.1 GL450828 EFN80310.1 KK852729 KDR17570.1 GBYB01007459 JAG77226.1 BT128196 KB632166 AEE63157.1 ERL89310.1 APGK01013754 APGK01023602 APGK01028435 KB740687 KB740505 KB739100 ENN79501.1 ENN80536.1 ENN82388.1 NNAY01000244 OXU29756.1 GEZM01021913 GEZM01021912 JAV88828.1 GECZ01017021 JAS52748.1 ADTU01011107 GL888932 EGI57318.1 KQ981673 KYN38229.1 GL767586 EFZ13421.1 KQ980581 KYN15414.1 LBMM01002445 KMQ94823.1 GEBQ01026659 JAT13318.1 GL435311 EFN73483.1 GECU01026418 JAS81288.1 PYGN01002498 PSN30380.1 DS235833 EEB18133.1 GEDC01026151 GEDC01012760 GEDC01004067 JAS11147.1 JAS24538.1 JAS33231.1 KQ977661 KYN00686.1 NEVH01016301 PNF25854.1 AJWK01028802 KQ434778 KZC04338.1 GFDF01003679 JAV10405.1 AJVK01059022 GFDL01008264 JAV26781.1 GFDL01008271 JAV26774.1 GFDL01007973 JAV27072.1 GANO01001163 JAB58708.1 JXUM01056738 JXUM01056739 JXUM01056740 JXUM01056741 JXUM01056742 KQ561923 KXJ77148.1 JXUM01076850 JXUM01076851 GAPW01003815 KQ562973 JAC09783.1 KXJ74728.1 QCYY01002621 ROT68915.1 GFDL01008053 JAV26992.1 DS232147 EDS36075.1 GFTR01003653 JAW12773.1 GECL01002047 JAP04077.1 GBGD01002866 JAC86023.1 JRES01000507 KNC30524.1 UFQT01000677 SSX26469.1 CM000157 EDW89560.1 CH954177 EDV58203.1 CH480816 EDW47161.1 CM000362 CM002911 EDX06402.1 KMY92585.1 AE013599 AY071695 AAF58892.1 AAL49317.1 AHN56053.1 KZ288192 PBC34282.1 JR042464 AEY59623.1 GDIQ01007246 JAN87491.1 GDIP01245558 JAI77843.1 JXJN01023092 GDRN01085609 JAI61329.1 CVRI01000036 CRK93103.1 KK120696 KFM78859.1 GDKW01001205 JAI55390.1 ACPB03019762 CCAG010017571 EZ422256 ADD18532.1 DS469615 EDO39013.1 LSMT01000012 PFX33440.1 AXCM01002007 GL732523 EFX90164.1 LJIJ01000157 ODN01353.1 CH933808 EDW09857.1 RCHS01002515 RMX46960.1 AAAB01008847 EAA06868.3 EGK96571.1 EGK96572.1 APCN01001050 GG666548 EEN56850.1 KK735465 KFR14173.1 JH432018 AXCN02001343 CH963846 EDW72553.1 QOIP01000004 RLU24168.1 CH940648 EDW61113.1 KRF79781.1 CH902619 EDV38100.1
AGBW02008449 OWR53338.1 KZ149987 PZC75667.1 NWSH01000440 PCG76412.1 ODYU01001638 SOQ38146.1 KQ459700 KPJ20392.1 GL450828 EFN80310.1 KK852729 KDR17570.1 GBYB01007459 JAG77226.1 BT128196 KB632166 AEE63157.1 ERL89310.1 APGK01013754 APGK01023602 APGK01028435 KB740687 KB740505 KB739100 ENN79501.1 ENN80536.1 ENN82388.1 NNAY01000244 OXU29756.1 GEZM01021913 GEZM01021912 JAV88828.1 GECZ01017021 JAS52748.1 ADTU01011107 GL888932 EGI57318.1 KQ981673 KYN38229.1 GL767586 EFZ13421.1 KQ980581 KYN15414.1 LBMM01002445 KMQ94823.1 GEBQ01026659 JAT13318.1 GL435311 EFN73483.1 GECU01026418 JAS81288.1 PYGN01002498 PSN30380.1 DS235833 EEB18133.1 GEDC01026151 GEDC01012760 GEDC01004067 JAS11147.1 JAS24538.1 JAS33231.1 KQ977661 KYN00686.1 NEVH01016301 PNF25854.1 AJWK01028802 KQ434778 KZC04338.1 GFDF01003679 JAV10405.1 AJVK01059022 GFDL01008264 JAV26781.1 GFDL01008271 JAV26774.1 GFDL01007973 JAV27072.1 GANO01001163 JAB58708.1 JXUM01056738 JXUM01056739 JXUM01056740 JXUM01056741 JXUM01056742 KQ561923 KXJ77148.1 JXUM01076850 JXUM01076851 GAPW01003815 KQ562973 JAC09783.1 KXJ74728.1 QCYY01002621 ROT68915.1 GFDL01008053 JAV26992.1 DS232147 EDS36075.1 GFTR01003653 JAW12773.1 GECL01002047 JAP04077.1 GBGD01002866 JAC86023.1 JRES01000507 KNC30524.1 UFQT01000677 SSX26469.1 CM000157 EDW89560.1 CH954177 EDV58203.1 CH480816 EDW47161.1 CM000362 CM002911 EDX06402.1 KMY92585.1 AE013599 AY071695 AAF58892.1 AAL49317.1 AHN56053.1 KZ288192 PBC34282.1 JR042464 AEY59623.1 GDIQ01007246 JAN87491.1 GDIP01245558 JAI77843.1 JXJN01023092 GDRN01085609 JAI61329.1 CVRI01000036 CRK93103.1 KK120696 KFM78859.1 GDKW01001205 JAI55390.1 ACPB03019762 CCAG010017571 EZ422256 ADD18532.1 DS469615 EDO39013.1 LSMT01000012 PFX33440.1 AXCM01002007 GL732523 EFX90164.1 LJIJ01000157 ODN01353.1 CH933808 EDW09857.1 RCHS01002515 RMX46960.1 AAAB01008847 EAA06868.3 EGK96571.1 EGK96572.1 APCN01001050 GG666548 EEN56850.1 KK735465 KFR14173.1 JH432018 AXCN02001343 CH963846 EDW72553.1 QOIP01000004 RLU24168.1 CH940648 EDW61113.1 KRF79781.1 CH902619 EDV38100.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000053240
UP000008237
+ More
UP000027135 UP000192223 UP000030742 UP000019118 UP000215335 UP000005205 UP000007755 UP000078541 UP000078492 UP000036403 UP000000311 UP000245037 UP000009046 UP000078542 UP000235965 UP000092461 UP000076502 UP000092462 UP000069940 UP000249989 UP000283509 UP000002320 UP000037069 UP000002282 UP000008711 UP000001292 UP000000304 UP000192221 UP000000803 UP000242457 UP000005203 UP000091820 UP000092443 UP000092460 UP000183832 UP000092445 UP000054359 UP000015103 UP000078200 UP000092444 UP000001593 UP000225706 UP000075883 UP000000305 UP000075920 UP000094527 UP000009192 UP000075885 UP000075902 UP000075901 UP000275408 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000095301 UP000001554 UP000053605 UP000075884 UP000075886 UP000002358 UP000085678 UP000007798 UP000075900 UP000279307 UP000008792 UP000007801
UP000027135 UP000192223 UP000030742 UP000019118 UP000215335 UP000005205 UP000007755 UP000078541 UP000078492 UP000036403 UP000000311 UP000245037 UP000009046 UP000078542 UP000235965 UP000092461 UP000076502 UP000092462 UP000069940 UP000249989 UP000283509 UP000002320 UP000037069 UP000002282 UP000008711 UP000001292 UP000000304 UP000192221 UP000000803 UP000242457 UP000005203 UP000091820 UP000092443 UP000092460 UP000183832 UP000092445 UP000054359 UP000015103 UP000078200 UP000092444 UP000001593 UP000225706 UP000075883 UP000000305 UP000075920 UP000094527 UP000009192 UP000075885 UP000075902 UP000075901 UP000275408 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000095301 UP000001554 UP000053605 UP000075884 UP000075886 UP000002358 UP000085678 UP000007798 UP000075900 UP000279307 UP000008792 UP000007801
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
H9IVH8
Q157R9
A0A194PDU7
A0A212FHX6
A0A2W1BR95
A0A2A4JYF3
+ More
A0A2H1VBH0 A0A194RSE0 E2BVD4 A0A067R381 A0A1W4XJM7 A0A0C9QUM8 J3JY72 N6UIR5 A0A232FG65 A0A1Y1MUI8 A0A1B6FRE5 A0A158NBH3 F4X8P9 A0A195FDX6 E9J188 A0A195DSB8 A0A0J7KWZ1 A0A1B6KPD5 E1ZZ68 A0A1B6I2Y0 A0A2P8XEF4 E0VXM7 A0A1B6E5M8 A0A151IGU3 A0A2J7QB94 A0A1B0GKF3 A0A154NZA9 A0A1L8DVT3 A0A1B0DFU5 A0A1Q3FGT9 A0A1Q3FGV5 A0A1Q3FHS5 U5EV63 A0A182GDX2 A0A023EKR7 A0A3R7Q583 A0A1Q3FHI9 B0WWI3 A0A224XX87 A0A0V0G840 A0A069DPN0 A0A0L0CGA7 A0A336MC22 B4NWL5 B3N6V4 B4HT51 B4QHR8 A0A1W4ULZ3 Q7JYX5 A0A2A3ESM8 V9IET7 A0A088AUK2 A0A0P6IEN8 A0A1A9W6F0 A0A0P4X8A1 A0A1A9YKG9 A0A1B0BZD1 A0A0P4VZ58 A0A1J1HYD7 A0A1A9ZHY5 A0A087UNC0 A0A0P4VTT1 T1IC92 A0A1A9USD0 D3TLF4 A7SBE8 A0A2B4SX42 A0A182ME07 E9FRH1 A0A182W671 A0A1D2N7V5 B4KMG9 A0A182PV13 A0A182TGI1 A0A182S9L1 A0A3M6TZW8 A0A182XY95 A0A182VN60 A0A182X7J7 F5HJV6 A0A182I9T9 A0A1I8MIC1 C3YRU5 A0A091WH27 T1JBQ6 A0A182MZX9 A0A182QZM5 K7IZA2 A0A1S3KBD9 B4MKB4 A0A182RYU6 A0A3L8DWL2 B4LNG1 B3MIM4
A0A2H1VBH0 A0A194RSE0 E2BVD4 A0A067R381 A0A1W4XJM7 A0A0C9QUM8 J3JY72 N6UIR5 A0A232FG65 A0A1Y1MUI8 A0A1B6FRE5 A0A158NBH3 F4X8P9 A0A195FDX6 E9J188 A0A195DSB8 A0A0J7KWZ1 A0A1B6KPD5 E1ZZ68 A0A1B6I2Y0 A0A2P8XEF4 E0VXM7 A0A1B6E5M8 A0A151IGU3 A0A2J7QB94 A0A1B0GKF3 A0A154NZA9 A0A1L8DVT3 A0A1B0DFU5 A0A1Q3FGT9 A0A1Q3FGV5 A0A1Q3FHS5 U5EV63 A0A182GDX2 A0A023EKR7 A0A3R7Q583 A0A1Q3FHI9 B0WWI3 A0A224XX87 A0A0V0G840 A0A069DPN0 A0A0L0CGA7 A0A336MC22 B4NWL5 B3N6V4 B4HT51 B4QHR8 A0A1W4ULZ3 Q7JYX5 A0A2A3ESM8 V9IET7 A0A088AUK2 A0A0P6IEN8 A0A1A9W6F0 A0A0P4X8A1 A0A1A9YKG9 A0A1B0BZD1 A0A0P4VZ58 A0A1J1HYD7 A0A1A9ZHY5 A0A087UNC0 A0A0P4VTT1 T1IC92 A0A1A9USD0 D3TLF4 A7SBE8 A0A2B4SX42 A0A182ME07 E9FRH1 A0A182W671 A0A1D2N7V5 B4KMG9 A0A182PV13 A0A182TGI1 A0A182S9L1 A0A3M6TZW8 A0A182XY95 A0A182VN60 A0A182X7J7 F5HJV6 A0A182I9T9 A0A1I8MIC1 C3YRU5 A0A091WH27 T1JBQ6 A0A182MZX9 A0A182QZM5 K7IZA2 A0A1S3KBD9 B4MKB4 A0A182RYU6 A0A3L8DWL2 B4LNG1 B3MIM4
PDB
4B93
E-value=1.0718e-61,
Score=597
Ontologies
GO
Topology
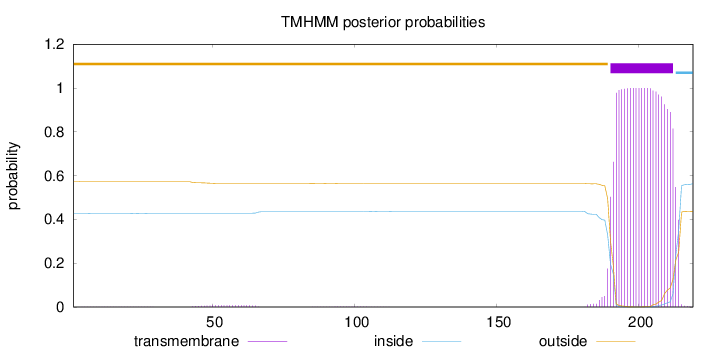
Length:
219
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.08671
Exp number, first 60 AAs:
0.13491
Total prob of N-in:
0.42757
outside
1 - 189
TMhelix
190 - 212
inside
213 - 219
Population Genetic Test Statistics
Pi
269.237191
Theta
175.365971
Tajima's D
0.823922
CLR
0.624537
CSRT
0.605669716514174
Interpretation
Uncertain
