Gene
KWMTBOMO07744
Pre Gene Modal
BGIBMGA001259
Annotation
PREDICTED:_otopetrin-2_isoform_X2_[Papilio_machaon]
Full name
Proton channel OtopLc
Alternative Name
Otopetrin-Lc
Location in the cell
PlasmaMembrane Reliability : 4.725
Sequence
CDS
ATGTCTGGTGGCTCTCATGTGTCGCGAACTACATTCGGCGGAGCCGACTCCGAGATGTCCAGTGGTAGTGCTCCACCGCTTCCCAGTCCAGCATCGGCTGGAGCCGCACTAGTCTCGCCAGAAACTTTAAGCAGACACAGCAGTCCGCCCCCGCTACCTCCTCATCCGTCGCACCAGCATCCGGCGCCATCTTGTTCTTCTCATCGGGTACATATGGAGTCGCCCTTACGGATGTCCAGTGCTCCATCAATGAACTTGGGTGGTTCATTGTCGCTACAACCCATGCTGTACGGAGCTGCGAGCGCTGGTTCGGCTCACGTTCCTCTGCCGCCAACAAGACGCTCACCACGACCTTCCAACACTATACAGCAGATGCACGTTATAAGCGCAAGCGATGCATTAGCGACCACGTTGTCCGCTCTCTACGGGAAGCTGTTGGTGGTCATGGGTGTCGCCTTTCCCATGGCCGAAGTCATATCAACGTACATTCCACCCTCGTTTTATGAGGGTTTCTATTTGTACCTGTACATCGGCAGCATGGTGTTCCTTTTGTATATGTACGCCGCTCTCGTAAGGGACAGAACGCGATCTGGAAAGGAAACCGCTATTGTTGCAAAGTCAGAATCTAGTTCGACGCCATCCGAATCGGATTCGTGTTCATCGCACGAACAAAGTACTACGACAACGACACACGCGATCCCACAGTTACCCGCCAAGAGATTATCCCTCGGATTTGCATTGGCCACCCGGACGCATCACTACGGAAGTTTTTATTTGCGAATGGGAGCTGTTGCTTTTGGTATCGGATCAATGATATACTCCGGGCTGGAATTTGGTCAGTACTGGGAACTGGAGAGGAACACTAATTGTCACAACGTCCTGCTGGCGCTGACACCCGCCACCAGAATGGCCTTCATCTTCATACAGATGTACTTTATCTTTCTCAACAACGAGATGAAAGTATACAAACACAAGATTGTGGCCAGGTTCGGTTTAATGCACATGGTTGGAACTAACCTCAGCGTGTGGTTGAATGTATTGGTTCAGGAAACGAAACACGAGATACTGACCTTCTACGACCCGGAAAATAAGACCATTGGACTCTCGCATAGACTAGCATTCCAAAAGGCCTTCGCTTCTTTAGCTACTGAGGTGCCTCCGTCACATCCGGCAGCGGCACATCTCCGCGTTCCGAGAGGGCTGAAAGGACCGCATCACATTTTCGAATGTAGACGTACTAACATAATGGGTACTCTCGTTCAAGATGCATCGCCTTTTCTGTTTCCTTGTACAATTGAGTATTCACTTATCTGTGCGGCTATACTTTACGTCATGTGGAAAAATATCTCGAAATATCCATCTAAAGACGTAGCGGCAGCAATCGCCAAAGCTCGCATACTCGAAGGCCTAGCTTACAAGAAGTCCCCACATTTGTACAGCGTCGACTGCGCTAGAGCTCACAAGGGACTGTTTTTTGGAATTCTCGTATTAGTTCTAACGATTATATCACTGATCTTGTTCTTCGTTTTAGTATCCAAAAAGGAATACGCATCATTGGCGGTTGTCGAAGTGAACATGTGTGAATTGGCTCTTTACGCTATGACGACTTTAGCTAGTTTGGCCGGAATGTTTTGTGTAAGAAATCTAAAGTATGACGGAAACCGAAATCTCGAACTCGATAACATACTTCTTGTTGGAGCTCAAACTGGCATGTTCATTTATGGTACGTTTACCATCATCGGAGGGCACTTCACTATAGAAAAGAACACGATTCTGATCTTGATCACTGCTTTATCTTCGTTAATCCAAACAACATGCCAGACGATGTTCATATTGGATGCGAGTCGTCGGTCGGCTAAGACTCCTGAACAATTGAGAAAAAAGCCTGGCAGGGAAATAGTGACGTTCTTATTGGTAACCAACTTAGCCATGTGGGCGATCAACACACTGGAGAAATCGCGCGCCGAATCTCACCCAGTCCAGTTGCACTTTTACGGACTCTGGGCTTGGACGATCATAACGCATGTCTCAATGCCGCTAGCGATTTTCTACAGATTCCACTCGACCGTTTGCTTGTGTGAGATTTGGAAGCGCGCTTACAAAATGAAACCTGCCTACATGTAG
Protein
MSGGSHVSRTTFGGADSEMSSGSAPPLPSPASAGAALVSPETLSRHSSPPPLPPHPSHQHPAPSCSSHRVHMESPLRMSSAPSMNLGGSLSLQPMLYGAASAGSAHVPLPPTRRSPRPSNTIQQMHVISASDALATTLSALYGKLLVVMGVAFPMAEVISTYIPPSFYEGFYLYLYIGSMVFLLYMYAALVRDRTRSGKETAIVAKSESSSTPSESDSCSSHEQSTTTTTHAIPQLPAKRLSLGFALATRTHHYGSFYLRMGAVAFGIGSMIYSGLEFGQYWELERNTNCHNVLLALTPATRMAFIFIQMYFIFLNNEMKVYKHKIVARFGLMHMVGTNLSVWLNVLVQETKHEILTFYDPENKTIGLSHRLAFQKAFASLATEVPPSHPAAAHLRVPRGLKGPHHIFECRRTNIMGTLVQDASPFLFPCTIEYSLICAAILYVMWKNISKYPSKDVAAAIAKARILEGLAYKKSPHLYSVDCARAHKGLFFGILVLVLTIISLILFFVLVSKKEYASLAVVEVNMCELALYAMTTLASLAGMFCVRNLKYDGNRNLELDNILLVGAQTGMFIYGTFTIIGGHFTIEKNTILILITALSSLIQTTCQTMFILDASRRSAKTPEQLRKKPGREIVTFLLVTNLAMWAINTLEKSRAESHPVQLHFYGLWAWTIITHVSMPLAIFYRFHSTVCLCEIWKRAYKMKPAYM
Summary
Description
Proton-selective channel that specifically transports protons into cells. Proton-selective channel activity is probably required in cell types that use changes in intracellular pH for cell signaling or to regulate biochemical or developmental processes.
Similarity
Belongs to the synaptobrevin family.
Belongs to the otopetrin family.
Belongs to the otopetrin family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Glycoprotein
Hydrogen ion transport
Ion channel
Ion transport
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Proton channel OtopLc
splice variant In isoform E.
splice variant In isoform E.
Uniprot
A0A2W1BL98
A0A194RSM4
A0A212FL14
A0A3S2NW75
H9IVH9
A0A2A4JFV7
+ More
D2A139 A0A2J7R2L2 A0A182QWY1 A0A0L7R7U6 A0A182RYU7 A0A0J7KQZ6 E2BVD7 A0A182MZX8 A0A182I9T7 A0A182UPD9 A0A182XY96 A0A182KK37 Q7QED6 A0A182PV14 A0A182MN77 A0A182X7J8 A0A084VGP7 F4X8Q0 E1ZZ69 A0A182TED6 A0A195DR75 A0A182W669 A0A088AUK5 V9IDZ6 A0A182SUD8 A0A182IW95 A0A182K9Q0 A0A182H3A5 A0A151IGX4 T1IC16 U4U5Z9 A0A158NBH1 A0A026W3F3 A0A1B6CS77 A0A1S4FER5 A0A1S4FEP3 A0A154NXD9 A0A1J1I405 A0A0K8UBX2 A0A232EUU7 A0A0K8VW96 Q174P9 A0A182FIF6 B4L407 K7IZA3 B0WWI4 A0A1W4VRK9 A0A0Q9WC09 A0A0R1ECV6 A0A0Q5T3I0 B7Z0W9-2 A0A1I8M6U8 A0A0M4EUV9 B7Z0W9 A0A3B0JDC8 A0A3B0JEK2 A0A146LN45 B7Z0W9-3 A0A1I8PUA9 A0A0P8XJU4 E0VXM8 A0A0C9S038 A0A0R3P453 A0A1A9X582 E9J190 W5JWD4 W8AWC5 A0A2R7WDC1 A0A0Q9WX90 A0A2S2NND9 A0A336KMM7 A0A3B0K0R2 X1X228 A0A1B0FL34 A0A1A9XRT5 A0A1A9UUQ5 A0A1B0AK69 A0A1B0AP93 B4I0M4 B4JLH0 B4R502 B4GWD2 J9JR25 A0A0L0BTM5 A0A2A3ETA9
D2A139 A0A2J7R2L2 A0A182QWY1 A0A0L7R7U6 A0A182RYU7 A0A0J7KQZ6 E2BVD7 A0A182MZX8 A0A182I9T7 A0A182UPD9 A0A182XY96 A0A182KK37 Q7QED6 A0A182PV14 A0A182MN77 A0A182X7J8 A0A084VGP7 F4X8Q0 E1ZZ69 A0A182TED6 A0A195DR75 A0A182W669 A0A088AUK5 V9IDZ6 A0A182SUD8 A0A182IW95 A0A182K9Q0 A0A182H3A5 A0A151IGX4 T1IC16 U4U5Z9 A0A158NBH1 A0A026W3F3 A0A1B6CS77 A0A1S4FER5 A0A1S4FEP3 A0A154NXD9 A0A1J1I405 A0A0K8UBX2 A0A232EUU7 A0A0K8VW96 Q174P9 A0A182FIF6 B4L407 K7IZA3 B0WWI4 A0A1W4VRK9 A0A0Q9WC09 A0A0R1ECV6 A0A0Q5T3I0 B7Z0W9-2 A0A1I8M6U8 A0A0M4EUV9 B7Z0W9 A0A3B0JDC8 A0A3B0JEK2 A0A146LN45 B7Z0W9-3 A0A1I8PUA9 A0A0P8XJU4 E0VXM8 A0A0C9S038 A0A0R3P453 A0A1A9X582 E9J190 W5JWD4 W8AWC5 A0A2R7WDC1 A0A0Q9WX90 A0A2S2NND9 A0A336KMM7 A0A3B0K0R2 X1X228 A0A1B0FL34 A0A1A9XRT5 A0A1A9UUQ5 A0A1B0AK69 A0A1B0AP93 B4I0M4 B4JLH0 B4R502 B4GWD2 J9JR25 A0A0L0BTM5 A0A2A3ETA9
Pubmed
28756777
26354079
22118469
19121390
18362917
19820115
+ More
20798317 25244985 20966253 12364791 24438588 21719571 26483478 23537049 21347285 24508170 30249741 17510324 28648823 17994087 20075255 17550304 10731132 12537572 29371428 25315136 26823975 20566863 15632085 21282665 20920257 23761445 24495485 26108605
20798317 25244985 20966253 12364791 24438588 21719571 26483478 23537049 21347285 24508170 30249741 17510324 28648823 17994087 20075255 17550304 10731132 12537572 29371428 25315136 26823975 20566863 15632085 21282665 20920257 23761445 24495485 26108605
EMBL
KZ149987
PZC75668.1
KQ459700
KPJ20394.1
AGBW02007913
OWR54427.1
+ More
RSAL01000049 RVE50377.1 BABH01001086 NWSH01001609 PCG70709.1 KQ971338 EFA02602.1 NEVH01007828 PNF35079.1 AXCN02001343 KQ414638 KOC66914.1 LBMM01004191 KMQ92716.1 GL450828 EFN80313.1 APCN01001049 AAAB01008847 EAA06870.5 AXCM01002007 ATLV01013047 KE524836 KFB37141.1 GL888932 EGI57319.1 GL435311 EFN73484.1 KQ980581 KYN15415.1 JR039392 JR039393 AEY58671.1 AEY58672.1 JXUM01024640 JXUM01024641 JXUM01024642 KQ560697 KXJ81233.1 KQ977661 KYN00685.1 ACPB03019762 KB632166 ERL89314.1 ADTU01011104 ADTU01011105 ADTU01011106 KK107453 QOIP01000004 EZA50595.1 RLU24169.1 GEDC01021123 JAS16175.1 KQ434778 KZC04336.1 CVRI01000036 CRK93105.1 GDHF01028243 JAI24071.1 NNAY01002084 OXU22132.1 GDHF01009509 JAI42805.1 CH477407 EAT41549.1 CH933810 EDW07285.2 DS232147 EDS36076.1 CH940651 KRF82211.1 CM000162 KRK05955.1 CH954180 KQS29759.1 AE014298 CP012528 ALC49626.1 OUUW01000003 SPP78613.1 SPP78612.1 GDHC01010417 JAQ08212.1 CH902622 KPU75104.1 DS235833 EEB18134.1 GBYB01013776 JAG83543.1 CH379063 KRT06293.1 GL767586 EFZ13432.1 ADMH02000170 ETN67515.1 GAMC01021459 GAMC01021458 JAB85096.1 KK854651 PTY17646.1 CH963851 KRF98206.1 GGMR01005657 MBY18276.1 UFQS01000677 UFQT01000677 SSX06116.1 SSX26472.1 SPP78611.1 ABLF02028005 ABLF02028012 CCAG010011710 JXJN01001277 JXJN01001278 JXJN01001279 CH480819 EDW53055.1 CH916370 EDW00423.1 CM000366 EDX17151.1 CH479194 EDW27016.1 ABLF02004943 ABLF02028010 ABLF02048961 JRES01001480 KNC22579.1 KZ288192 PBC34281.1
RSAL01000049 RVE50377.1 BABH01001086 NWSH01001609 PCG70709.1 KQ971338 EFA02602.1 NEVH01007828 PNF35079.1 AXCN02001343 KQ414638 KOC66914.1 LBMM01004191 KMQ92716.1 GL450828 EFN80313.1 APCN01001049 AAAB01008847 EAA06870.5 AXCM01002007 ATLV01013047 KE524836 KFB37141.1 GL888932 EGI57319.1 GL435311 EFN73484.1 KQ980581 KYN15415.1 JR039392 JR039393 AEY58671.1 AEY58672.1 JXUM01024640 JXUM01024641 JXUM01024642 KQ560697 KXJ81233.1 KQ977661 KYN00685.1 ACPB03019762 KB632166 ERL89314.1 ADTU01011104 ADTU01011105 ADTU01011106 KK107453 QOIP01000004 EZA50595.1 RLU24169.1 GEDC01021123 JAS16175.1 KQ434778 KZC04336.1 CVRI01000036 CRK93105.1 GDHF01028243 JAI24071.1 NNAY01002084 OXU22132.1 GDHF01009509 JAI42805.1 CH477407 EAT41549.1 CH933810 EDW07285.2 DS232147 EDS36076.1 CH940651 KRF82211.1 CM000162 KRK05955.1 CH954180 KQS29759.1 AE014298 CP012528 ALC49626.1 OUUW01000003 SPP78613.1 SPP78612.1 GDHC01010417 JAQ08212.1 CH902622 KPU75104.1 DS235833 EEB18134.1 GBYB01013776 JAG83543.1 CH379063 KRT06293.1 GL767586 EFZ13432.1 ADMH02000170 ETN67515.1 GAMC01021459 GAMC01021458 JAB85096.1 KK854651 PTY17646.1 CH963851 KRF98206.1 GGMR01005657 MBY18276.1 UFQS01000677 UFQT01000677 SSX06116.1 SSX26472.1 SPP78611.1 ABLF02028005 ABLF02028012 CCAG010011710 JXJN01001277 JXJN01001278 JXJN01001279 CH480819 EDW53055.1 CH916370 EDW00423.1 CM000366 EDX17151.1 CH479194 EDW27016.1 ABLF02004943 ABLF02028010 ABLF02048961 JRES01001480 KNC22579.1 KZ288192 PBC34281.1
Proteomes
UP000053240
UP000007151
UP000283053
UP000005204
UP000218220
UP000007266
+ More
UP000235965 UP000075886 UP000053825 UP000075900 UP000036403 UP000008237 UP000075884 UP000075840 UP000075903 UP000076408 UP000075882 UP000007062 UP000075885 UP000075883 UP000076407 UP000030765 UP000007755 UP000000311 UP000075902 UP000078492 UP000075920 UP000005203 UP000075901 UP000075880 UP000075881 UP000069940 UP000249989 UP000078542 UP000015103 UP000030742 UP000005205 UP000053097 UP000279307 UP000076502 UP000183832 UP000215335 UP000008820 UP000069272 UP000009192 UP000002358 UP000002320 UP000192221 UP000008792 UP000002282 UP000008711 UP000000803 UP000095301 UP000092553 UP000268350 UP000095300 UP000007801 UP000009046 UP000001819 UP000091820 UP000000673 UP000007798 UP000007819 UP000092444 UP000092443 UP000078200 UP000092445 UP000092460 UP000001292 UP000001070 UP000000304 UP000008744 UP000037069 UP000242457
UP000235965 UP000075886 UP000053825 UP000075900 UP000036403 UP000008237 UP000075884 UP000075840 UP000075903 UP000076408 UP000075882 UP000007062 UP000075885 UP000075883 UP000076407 UP000030765 UP000007755 UP000000311 UP000075902 UP000078492 UP000075920 UP000005203 UP000075901 UP000075880 UP000075881 UP000069940 UP000249989 UP000078542 UP000015103 UP000030742 UP000005205 UP000053097 UP000279307 UP000076502 UP000183832 UP000215335 UP000008820 UP000069272 UP000009192 UP000002358 UP000002320 UP000192221 UP000008792 UP000002282 UP000008711 UP000000803 UP000095301 UP000092553 UP000268350 UP000095300 UP000007801 UP000009046 UP000001819 UP000091820 UP000000673 UP000007798 UP000007819 UP000092444 UP000092443 UP000078200 UP000092445 UP000092460 UP000001292 UP000001070 UP000000304 UP000008744 UP000037069 UP000242457
Interpro
SUPFAM
SSF64356
SSF64356
ProteinModelPortal
A0A2W1BL98
A0A194RSM4
A0A212FL14
A0A3S2NW75
H9IVH9
A0A2A4JFV7
+ More
D2A139 A0A2J7R2L2 A0A182QWY1 A0A0L7R7U6 A0A182RYU7 A0A0J7KQZ6 E2BVD7 A0A182MZX8 A0A182I9T7 A0A182UPD9 A0A182XY96 A0A182KK37 Q7QED6 A0A182PV14 A0A182MN77 A0A182X7J8 A0A084VGP7 F4X8Q0 E1ZZ69 A0A182TED6 A0A195DR75 A0A182W669 A0A088AUK5 V9IDZ6 A0A182SUD8 A0A182IW95 A0A182K9Q0 A0A182H3A5 A0A151IGX4 T1IC16 U4U5Z9 A0A158NBH1 A0A026W3F3 A0A1B6CS77 A0A1S4FER5 A0A1S4FEP3 A0A154NXD9 A0A1J1I405 A0A0K8UBX2 A0A232EUU7 A0A0K8VW96 Q174P9 A0A182FIF6 B4L407 K7IZA3 B0WWI4 A0A1W4VRK9 A0A0Q9WC09 A0A0R1ECV6 A0A0Q5T3I0 B7Z0W9-2 A0A1I8M6U8 A0A0M4EUV9 B7Z0W9 A0A3B0JDC8 A0A3B0JEK2 A0A146LN45 B7Z0W9-3 A0A1I8PUA9 A0A0P8XJU4 E0VXM8 A0A0C9S038 A0A0R3P453 A0A1A9X582 E9J190 W5JWD4 W8AWC5 A0A2R7WDC1 A0A0Q9WX90 A0A2S2NND9 A0A336KMM7 A0A3B0K0R2 X1X228 A0A1B0FL34 A0A1A9XRT5 A0A1A9UUQ5 A0A1B0AK69 A0A1B0AP93 B4I0M4 B4JLH0 B4R502 B4GWD2 J9JR25 A0A0L0BTM5 A0A2A3ETA9
D2A139 A0A2J7R2L2 A0A182QWY1 A0A0L7R7U6 A0A182RYU7 A0A0J7KQZ6 E2BVD7 A0A182MZX8 A0A182I9T7 A0A182UPD9 A0A182XY96 A0A182KK37 Q7QED6 A0A182PV14 A0A182MN77 A0A182X7J8 A0A084VGP7 F4X8Q0 E1ZZ69 A0A182TED6 A0A195DR75 A0A182W669 A0A088AUK5 V9IDZ6 A0A182SUD8 A0A182IW95 A0A182K9Q0 A0A182H3A5 A0A151IGX4 T1IC16 U4U5Z9 A0A158NBH1 A0A026W3F3 A0A1B6CS77 A0A1S4FER5 A0A1S4FEP3 A0A154NXD9 A0A1J1I405 A0A0K8UBX2 A0A232EUU7 A0A0K8VW96 Q174P9 A0A182FIF6 B4L407 K7IZA3 B0WWI4 A0A1W4VRK9 A0A0Q9WC09 A0A0R1ECV6 A0A0Q5T3I0 B7Z0W9-2 A0A1I8M6U8 A0A0M4EUV9 B7Z0W9 A0A3B0JDC8 A0A3B0JEK2 A0A146LN45 B7Z0W9-3 A0A1I8PUA9 A0A0P8XJU4 E0VXM8 A0A0C9S038 A0A0R3P453 A0A1A9X582 E9J190 W5JWD4 W8AWC5 A0A2R7WDC1 A0A0Q9WX90 A0A2S2NND9 A0A336KMM7 A0A3B0K0R2 X1X228 A0A1B0FL34 A0A1A9XRT5 A0A1A9UUQ5 A0A1B0AK69 A0A1B0AP93 B4I0M4 B4JLH0 B4R502 B4GWD2 J9JR25 A0A0L0BTM5 A0A2A3ETA9
PDB
6NF6
E-value=5.17519e-14,
Score=191
Ontologies
PANTHER
Topology
Subcellular location
Cell membrane
Length:
707
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
259.45628
Exp number, first 60 AAs:
0.00177
Total prob of N-in:
0.97719
inside
1 - 133
TMhelix
134 - 156
outside
157 - 170
TMhelix
171 - 191
inside
192 - 255
TMhelix
256 - 278
outside
279 - 292
TMhelix
293 - 315
inside
316 - 326
TMhelix
327 - 349
outside
350 - 423
TMhelix
424 - 446
inside
447 - 488
TMhelix
489 - 511
outside
512 - 525
TMhelix
526 - 548
inside
549 - 560
TMhelix
561 - 580
outside
581 - 589
TMhelix
590 - 612
inside
613 - 632
TMhelix
633 - 650
outside
651 - 664
TMhelix
665 - 687
inside
688 - 707
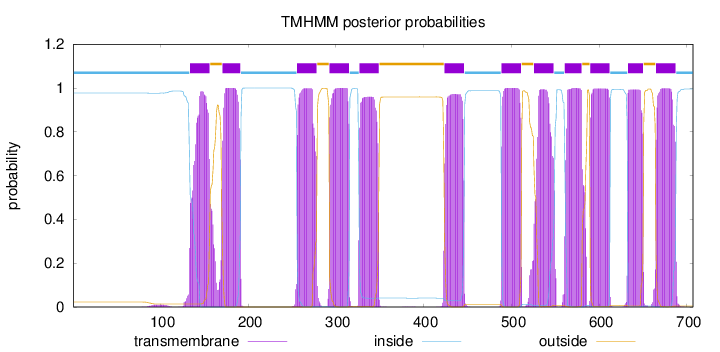
Population Genetic Test Statistics
Pi
226.36172
Theta
176.802984
Tajima's D
0.847423
CLR
0.457882
CSRT
0.616819159042048
Interpretation
Uncertain
