Gene
KWMTBOMO07727
Pre Gene Modal
BGIBMGA000882
Annotation
39S_ribosomal_protein_L27?_mitochondrial_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.883 Mitochondrial Reliability : 1.887
Sequence
CDS
ATGTCATTTAACATTTTACTCAAAACATCAGAAAGAGGTATTTTTCTAAAAGAGCTTGTCCGGAATGCCAGTAAAAAAACCGGAGGTAGTACGCAGAATACTAATTGTAAAGTAAAACCAAAGCACAGAGGGTGGAAAGTACAAGATGGGCATTTTGTTCAAGCGGGACATATGTTAGCTACGCAAAGAACCACTCGGTTTCATCCAGGATTGAATGTTGGTTTTGGTGTTAATGGTACTTTGTTTGCAATGGAGGCTGGAAAAGTAGTTGTCACATGTGAAAAGTTTGATCCAAACTGGGATCACACTTGGGTCCAGAGGATGTACAAAGGGCGCTATGACCAGACCATCTATAAAAAATATTACAATGTTATACCAGAGCCCCAACATCAGAGATTTAAACTTATTGATGAAGTGTAA
Protein
MSFNILLKTSERGIFLKELVRNASKKTGGSTQNTNCKVKPKHRGWKVQDGHFVQAGHMLATQRTTRFHPGLNVGFGVNGTLFAMEAGKVVVTCEKFDPNWDHTWVQRMYKGRYDQTIYKKYYNVIPEPQHQRFKLIDEV
Summary
Uniprot
Q2F676
H9IUF2
A0A2A4JV64
A0A2H1VGX0
A0A0L7KRB2
A0A194RSN5
+ More
A0A194PD68 A0A212FAH3 A0A182QEJ4 A0A182JRI7 A0A182R364 A0A182JFB0 A0A182NLB0 A0A182XWW3 B4NYY1 B3N3T1 A0A182XI08 A0A182TR95 A0A0K8TRR1 A0A182VCK0 A0A182HSY9 A0A182VXE3 A0A1W4WAL8 A0A0K8VHT6 A0A034V099 Q7QC64 A0A182PAF2 A0A0A1WFY0 A0A2M4AVS6 B3MU24 A0A2M4C2V1 A0A2M4C2U9 W8CEE1 B4LUW9 A0A182FT48 A0A0K8VS69 B4KLF6 A0A023EER1 A0A1A9VW29 A0A1B2AJA3 W5JM70 B4QA81 B4I399 A0A1A9XJN9 B4MZL4 A0A2M3ZCX9 B5RJ05 A0A0L0BSG7 Q29LK6 A0A1B0C305 A0A182SHR0 A0A1S4F1P0 A0A0M5IVX0 Q17I88 B4GPT5 Q86BM8 B0X4E4 A0A1Q3FJ80 A0A1I8MIH3 A0A1I8Q5F4 A0A1A9WKP2 B4JQS2 A0A0J7NW46 D6WIR5 A0A2S2R7E1 C4WXZ8 N6TD57 K7IQN7 R4V3V8 A0A232EU87 U4U6Z1 A0A2H8TIT5 A0A1W4XT03 A0A2P8XWX4 A0A067R3P3 A0A1B0CDW4 T1HG42 A0A0P4VLB3 A0A0L7QPE9 A0A026VYV4 A0A310SM28 A0A1L8E187 A0A0V0GAI5 A0A224XPI0 F4WKV1 A0A195BJH2 A0A158P3X1 A0A2A3E217 A0A151JVB5 E2AUD6 A0A154PDH4 A0A1Y1L7J6 A0A088AW11 A0A1J1I3V2 A0A1B6I9C8 A0A1B6GSW5 A0A151J8S6 A0A1B6L6Y0 A0A1B6CC52 A0A0A9YXG4
A0A194PD68 A0A212FAH3 A0A182QEJ4 A0A182JRI7 A0A182R364 A0A182JFB0 A0A182NLB0 A0A182XWW3 B4NYY1 B3N3T1 A0A182XI08 A0A182TR95 A0A0K8TRR1 A0A182VCK0 A0A182HSY9 A0A182VXE3 A0A1W4WAL8 A0A0K8VHT6 A0A034V099 Q7QC64 A0A182PAF2 A0A0A1WFY0 A0A2M4AVS6 B3MU24 A0A2M4C2V1 A0A2M4C2U9 W8CEE1 B4LUW9 A0A182FT48 A0A0K8VS69 B4KLF6 A0A023EER1 A0A1A9VW29 A0A1B2AJA3 W5JM70 B4QA81 B4I399 A0A1A9XJN9 B4MZL4 A0A2M3ZCX9 B5RJ05 A0A0L0BSG7 Q29LK6 A0A1B0C305 A0A182SHR0 A0A1S4F1P0 A0A0M5IVX0 Q17I88 B4GPT5 Q86BM8 B0X4E4 A0A1Q3FJ80 A0A1I8MIH3 A0A1I8Q5F4 A0A1A9WKP2 B4JQS2 A0A0J7NW46 D6WIR5 A0A2S2R7E1 C4WXZ8 N6TD57 K7IQN7 R4V3V8 A0A232EU87 U4U6Z1 A0A2H8TIT5 A0A1W4XT03 A0A2P8XWX4 A0A067R3P3 A0A1B0CDW4 T1HG42 A0A0P4VLB3 A0A0L7QPE9 A0A026VYV4 A0A310SM28 A0A1L8E187 A0A0V0GAI5 A0A224XPI0 F4WKV1 A0A195BJH2 A0A158P3X1 A0A2A3E217 A0A151JVB5 E2AUD6 A0A154PDH4 A0A1Y1L7J6 A0A088AW11 A0A1J1I3V2 A0A1B6I9C8 A0A1B6GSW5 A0A151J8S6 A0A1B6L6Y0 A0A1B6CC52 A0A0A9YXG4
Pubmed
19121390
26227816
26354079
22118469
25244985
17994087
+ More
17550304 26369729 25348373 12364791 25830018 24495485 24945155 26483478 20920257 23761445 22936249 26108605 15632085 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18362917 19820115 23537049 20075255 28648823 29403074 24845553 27129103 24508170 30249741 21719571 21347285 20798317 28004739 25401762 26823975
17550304 26369729 25348373 12364791 25830018 24495485 24945155 26483478 20920257 23761445 22936249 26108605 15632085 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18362917 19820115 23537049 20075255 28648823 29403074 24845553 27129103 24508170 30249741 21719571 21347285 20798317 28004739 25401762 26823975
EMBL
DQ311196
ABD36141.2
BABH01001167
NWSH01000518
PCG75901.1
ODYU01002500
+ More
SOQ40057.1 JTDY01006620 KOB65832.1 KQ459700 KPJ20404.1 KQ459606 KPI91182.1 AGBW02009477 OWR50727.1 AXCN02000116 CM000157 EDW87645.1 CH954177 EDV57740.2 GDAI01000551 JAI17052.1 APCN01000376 GDHF01014199 JAI38115.1 GAKP01023386 JAC35572.1 AAAB01008859 EAA08179.5 GBXI01016363 JAC97928.1 GGFK01011407 MBW44728.1 CH902624 EDV33353.1 GGFJ01010492 MBW59633.1 GGFJ01010491 MBW59632.1 GAMC01000356 JAC06200.1 CH940649 EDW64296.1 GDHF01010593 JAI41721.1 CH933807 EDW12837.2 JXUM01102803 GAPW01005731 KQ564753 JAC07867.1 KXJ71792.1 KX531421 ANY27231.1 ADMH02001133 ETN63995.1 CM000361 CM002910 EDX03719.1 KMY88088.1 CH480820 EDW54244.1 CH963920 EDW77799.1 GGFM01005610 MBW26361.1 BT044279 ACH92344.1 JRES01001441 KNC22918.1 CH379060 EAL34039.3 JXJN01024742 CP012523 ALC38111.1 CH477241 EAT46409.1 CH479187 EDW39607.1 AE014134 AAO41158.1 AGB92564.1 AHN54117.1 DS232338 EDS40297.1 GFDL01007509 JAV27536.1 CH916372 EDV99252.1 LBMM01001275 KMQ96600.1 KQ971321 EEZ99502.1 GGMS01016736 MBY85939.1 ABLF02023377 AK342943 BAH72768.1 APGK01042723 KB741007 ENN75633.1 KC740674 AGM32498.1 NNAY01002176 OXU21866.1 KB631811 ERL86351.1 GFXV01002116 MBW13921.1 PYGN01001224 PSN36511.1 KK852726 KDR17653.1 AJWK01008307 ACPB03001139 GDKW01003402 JAI53193.1 KQ414815 KOC60518.1 KK107570 QOIP01000005 EZA48855.1 RLU22610.1 KQ764027 OAD54629.1 GFDF01001678 JAV12406.1 GECL01000972 JAP05152.1 GFTR01002030 JAW14396.1 GL888206 EGI65193.1 KQ976453 KYM85323.1 ADTU01008608 KZ288451 PBC25574.1 KQ981701 KYN37478.1 GL442815 EFN62993.1 KQ434878 KZC09902.1 GEZM01062527 GEZM01062524 GEZM01062523 GEZM01062521 GEZM01062520 GEZM01062519 GEZM01062518 GEZM01062516 JAV69659.1 CVRI01000040 CRK94875.1 GECU01024187 JAS83519.1 GECZ01004239 JAS65530.1 KQ979480 KYN21263.1 GEBQ01020494 JAT19483.1 GEDC01026264 JAS11034.1 GBHO01009354 GBRD01007495 GDHC01017588 JAG34250.1 JAG58326.1 JAQ01041.1
SOQ40057.1 JTDY01006620 KOB65832.1 KQ459700 KPJ20404.1 KQ459606 KPI91182.1 AGBW02009477 OWR50727.1 AXCN02000116 CM000157 EDW87645.1 CH954177 EDV57740.2 GDAI01000551 JAI17052.1 APCN01000376 GDHF01014199 JAI38115.1 GAKP01023386 JAC35572.1 AAAB01008859 EAA08179.5 GBXI01016363 JAC97928.1 GGFK01011407 MBW44728.1 CH902624 EDV33353.1 GGFJ01010492 MBW59633.1 GGFJ01010491 MBW59632.1 GAMC01000356 JAC06200.1 CH940649 EDW64296.1 GDHF01010593 JAI41721.1 CH933807 EDW12837.2 JXUM01102803 GAPW01005731 KQ564753 JAC07867.1 KXJ71792.1 KX531421 ANY27231.1 ADMH02001133 ETN63995.1 CM000361 CM002910 EDX03719.1 KMY88088.1 CH480820 EDW54244.1 CH963920 EDW77799.1 GGFM01005610 MBW26361.1 BT044279 ACH92344.1 JRES01001441 KNC22918.1 CH379060 EAL34039.3 JXJN01024742 CP012523 ALC38111.1 CH477241 EAT46409.1 CH479187 EDW39607.1 AE014134 AAO41158.1 AGB92564.1 AHN54117.1 DS232338 EDS40297.1 GFDL01007509 JAV27536.1 CH916372 EDV99252.1 LBMM01001275 KMQ96600.1 KQ971321 EEZ99502.1 GGMS01016736 MBY85939.1 ABLF02023377 AK342943 BAH72768.1 APGK01042723 KB741007 ENN75633.1 KC740674 AGM32498.1 NNAY01002176 OXU21866.1 KB631811 ERL86351.1 GFXV01002116 MBW13921.1 PYGN01001224 PSN36511.1 KK852726 KDR17653.1 AJWK01008307 ACPB03001139 GDKW01003402 JAI53193.1 KQ414815 KOC60518.1 KK107570 QOIP01000005 EZA48855.1 RLU22610.1 KQ764027 OAD54629.1 GFDF01001678 JAV12406.1 GECL01000972 JAP05152.1 GFTR01002030 JAW14396.1 GL888206 EGI65193.1 KQ976453 KYM85323.1 ADTU01008608 KZ288451 PBC25574.1 KQ981701 KYN37478.1 GL442815 EFN62993.1 KQ434878 KZC09902.1 GEZM01062527 GEZM01062524 GEZM01062523 GEZM01062521 GEZM01062520 GEZM01062519 GEZM01062518 GEZM01062516 JAV69659.1 CVRI01000040 CRK94875.1 GECU01024187 JAS83519.1 GECZ01004239 JAS65530.1 KQ979480 KYN21263.1 GEBQ01020494 JAT19483.1 GEDC01026264 JAS11034.1 GBHO01009354 GBRD01007495 GDHC01017588 JAG34250.1 JAG58326.1 JAQ01041.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000075886 UP000075881 UP000075900 UP000075880 UP000075884 UP000076408 UP000002282 UP000008711 UP000076407 UP000075902 UP000075903 UP000075840 UP000075920 UP000192221 UP000007062 UP000075885 UP000007801 UP000008792 UP000069272 UP000009192 UP000069940 UP000249989 UP000078200 UP000000673 UP000000304 UP000001292 UP000092443 UP000007798 UP000037069 UP000001819 UP000092460 UP000075901 UP000092553 UP000008820 UP000008744 UP000000803 UP000002320 UP000095301 UP000095300 UP000091820 UP000001070 UP000036403 UP000007266 UP000007819 UP000019118 UP000002358 UP000215335 UP000030742 UP000192223 UP000245037 UP000027135 UP000092461 UP000015103 UP000053825 UP000053097 UP000279307 UP000007755 UP000078540 UP000005205 UP000242457 UP000078541 UP000000311 UP000076502 UP000005203 UP000183832 UP000078492
UP000075886 UP000075881 UP000075900 UP000075880 UP000075884 UP000076408 UP000002282 UP000008711 UP000076407 UP000075902 UP000075903 UP000075840 UP000075920 UP000192221 UP000007062 UP000075885 UP000007801 UP000008792 UP000069272 UP000009192 UP000069940 UP000249989 UP000078200 UP000000673 UP000000304 UP000001292 UP000092443 UP000007798 UP000037069 UP000001819 UP000092460 UP000075901 UP000092553 UP000008820 UP000008744 UP000000803 UP000002320 UP000095301 UP000095300 UP000091820 UP000001070 UP000036403 UP000007266 UP000007819 UP000019118 UP000002358 UP000215335 UP000030742 UP000192223 UP000245037 UP000027135 UP000092461 UP000015103 UP000053825 UP000053097 UP000279307 UP000007755 UP000078540 UP000005205 UP000242457 UP000078541 UP000000311 UP000076502 UP000005203 UP000183832 UP000078492
Pfam
PF01016 Ribosomal_L27
Interpro
IPR001684
Ribosomal_L27
ProteinModelPortal
Q2F676
H9IUF2
A0A2A4JV64
A0A2H1VGX0
A0A0L7KRB2
A0A194RSN5
+ More
A0A194PD68 A0A212FAH3 A0A182QEJ4 A0A182JRI7 A0A182R364 A0A182JFB0 A0A182NLB0 A0A182XWW3 B4NYY1 B3N3T1 A0A182XI08 A0A182TR95 A0A0K8TRR1 A0A182VCK0 A0A182HSY9 A0A182VXE3 A0A1W4WAL8 A0A0K8VHT6 A0A034V099 Q7QC64 A0A182PAF2 A0A0A1WFY0 A0A2M4AVS6 B3MU24 A0A2M4C2V1 A0A2M4C2U9 W8CEE1 B4LUW9 A0A182FT48 A0A0K8VS69 B4KLF6 A0A023EER1 A0A1A9VW29 A0A1B2AJA3 W5JM70 B4QA81 B4I399 A0A1A9XJN9 B4MZL4 A0A2M3ZCX9 B5RJ05 A0A0L0BSG7 Q29LK6 A0A1B0C305 A0A182SHR0 A0A1S4F1P0 A0A0M5IVX0 Q17I88 B4GPT5 Q86BM8 B0X4E4 A0A1Q3FJ80 A0A1I8MIH3 A0A1I8Q5F4 A0A1A9WKP2 B4JQS2 A0A0J7NW46 D6WIR5 A0A2S2R7E1 C4WXZ8 N6TD57 K7IQN7 R4V3V8 A0A232EU87 U4U6Z1 A0A2H8TIT5 A0A1W4XT03 A0A2P8XWX4 A0A067R3P3 A0A1B0CDW4 T1HG42 A0A0P4VLB3 A0A0L7QPE9 A0A026VYV4 A0A310SM28 A0A1L8E187 A0A0V0GAI5 A0A224XPI0 F4WKV1 A0A195BJH2 A0A158P3X1 A0A2A3E217 A0A151JVB5 E2AUD6 A0A154PDH4 A0A1Y1L7J6 A0A088AW11 A0A1J1I3V2 A0A1B6I9C8 A0A1B6GSW5 A0A151J8S6 A0A1B6L6Y0 A0A1B6CC52 A0A0A9YXG4
A0A194PD68 A0A212FAH3 A0A182QEJ4 A0A182JRI7 A0A182R364 A0A182JFB0 A0A182NLB0 A0A182XWW3 B4NYY1 B3N3T1 A0A182XI08 A0A182TR95 A0A0K8TRR1 A0A182VCK0 A0A182HSY9 A0A182VXE3 A0A1W4WAL8 A0A0K8VHT6 A0A034V099 Q7QC64 A0A182PAF2 A0A0A1WFY0 A0A2M4AVS6 B3MU24 A0A2M4C2V1 A0A2M4C2U9 W8CEE1 B4LUW9 A0A182FT48 A0A0K8VS69 B4KLF6 A0A023EER1 A0A1A9VW29 A0A1B2AJA3 W5JM70 B4QA81 B4I399 A0A1A9XJN9 B4MZL4 A0A2M3ZCX9 B5RJ05 A0A0L0BSG7 Q29LK6 A0A1B0C305 A0A182SHR0 A0A1S4F1P0 A0A0M5IVX0 Q17I88 B4GPT5 Q86BM8 B0X4E4 A0A1Q3FJ80 A0A1I8MIH3 A0A1I8Q5F4 A0A1A9WKP2 B4JQS2 A0A0J7NW46 D6WIR5 A0A2S2R7E1 C4WXZ8 N6TD57 K7IQN7 R4V3V8 A0A232EU87 U4U6Z1 A0A2H8TIT5 A0A1W4XT03 A0A2P8XWX4 A0A067R3P3 A0A1B0CDW4 T1HG42 A0A0P4VLB3 A0A0L7QPE9 A0A026VYV4 A0A310SM28 A0A1L8E187 A0A0V0GAI5 A0A224XPI0 F4WKV1 A0A195BJH2 A0A158P3X1 A0A2A3E217 A0A151JVB5 E2AUD6 A0A154PDH4 A0A1Y1L7J6 A0A088AW11 A0A1J1I3V2 A0A1B6I9C8 A0A1B6GSW5 A0A151J8S6 A0A1B6L6Y0 A0A1B6CC52 A0A0A9YXG4
PDB
6GB2
E-value=1.68666e-18,
Score=221
Ontologies
GO
PANTHER
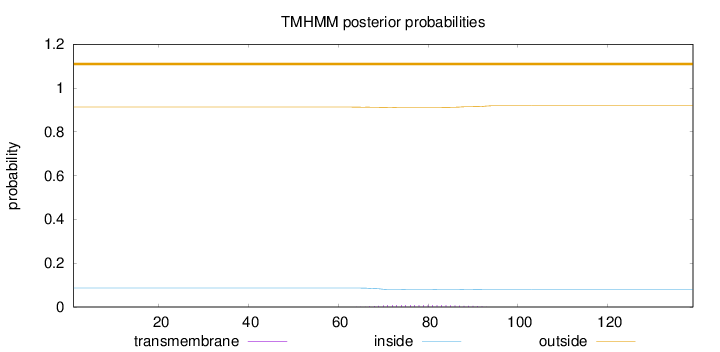
Topology
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19167
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.08686
outside
1 - 139
Population Genetic Test Statistics
Pi
335.5045
Theta
229.168807
Tajima's D
1.461268
CLR
0
CSRT
0.778311084445778
Interpretation
Uncertain
