Gene
KWMTBOMO07726
Pre Gene Modal
BGIBMGA001264
Annotation
PREDICTED:_transcriptional_adapter_1-like_[Amyelois_transitella]
Full name
Transcriptional adapter 1
Alternative Name
Transcriptional adapter 1-like protein
Location in the cell
Cytoplasmic Reliability : 1.848
Sequence
CDS
ATGGCTTCCGATACATTAAATATAGCACGGCGAAAATTAAATGAAGTACTCGGTGAAAAGTCAACAAAGTATTTCAATCAAATGAAATTGTGGTTCCGTATGAAACTGACTAAGGAAGAATTCGACGGAGAAGCAAGGAAACTATTAAGTAACGACCAAGTACATTTCCATAACGAGTTTTTGTTAGCATTATTGAACAAAGTTGAAGGCTTAGCAGAAACTTCGATTACAATAGCCCAAGAGAAAGCTAATTCACACAACAGAAACAGTCGGAGACATAAGAGAAGCTCTAGAACCTCTGAGAAGAGTAACTTTGAACCAGTTGATATACTTGAGTATCTGCCGCCTAATTCACCACCTGGAGCTGGCAGTGACGGTGTCAAATATGCAACACAAGAAATATTTCTACCTGATCATGCTCTAGTTGTGGGCAGATTCATGTTGGCCGCCTGGGAACTTGGCTTAGAAGGAGCGGATGACGATGCTGCTGACATTGTAGTGGTAGCTGTACAGAATTTCCTCAAAAATGTAATAAGTGCTGTTTTAGCACAAAGGAAAGGCTACAAAACTCATGGGAAGTACTTCATGTACGATATTGGTGGTGACATGCCGAATATGTGGCTGAGAAACTCAGCCAAACTATACGATCCTCAGAGTTGGGGAAGGGTAAATGTAGATGATGGAGTGGATTCCCTCGGTCCCAGATGTCCTCCTACAATTGATGAAATGGAACATTCAGCTGTTTTTGAAATTGCCTGCAGTGCCCCAAATCCAGAACCAAATGAAGACAGACTAACAATAGATGAATTCTACAATACCTTATTGACACACAGAAATATAATTGCATGTCATTCAATCTATGCAATCAATATGGAGAGGTTATCGGTCATGTTAAATCATCCTAGTTATTAG
Protein
MASDTLNIARRKLNEVLGEKSTKYFNQMKLWFRMKLTKEEFDGEARKLLSNDQVHFHNEFLLALLNKVEGLAETSITIAQEKANSHNRNSRRHKRSSRTSEKSNFEPVDILEYLPPNSPPGAGSDGVKYATQEIFLPDHALVVGRFMLAAWELGLEGADDDAADIVVVAVQNFLKNVISAVLAQRKGYKTHGKYFMYDIGGDMPNMWLRNSAKLYDPQSWGRVNVDDGVDSLGPRCPPTIDEMEHSAVFEIACSAPNPEPNEDRLTIDEFYNTLLTHRNIIACHSIYAINMERLSVMLNHPSY
Summary
Similarity
Belongs to the TADA1 family.
Keywords
Complete proteome
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Transcriptional adapter 1
Uniprot
A0A194RW40
A0A3S2TD59
A0A1E1WKL1
A0A2A4JWW4
A0A2H1VGV9
A0A212EY78
+ More
H9IVI4 I4DPA6 A0A194PJB7 A0A2W1BRI2 S4NR67 A0A0L7K3C9 A0A067QY89 A0A2J7PC59 E0VAQ9 A0A0L7RG24 A0A0M8ZRT2 A0A2A3E9Q6 A0A088A274 A0A310SNF1 A0A154NW62 A0A0C9RAW3 E2BA55 F4WWC6 A0A195FAZ0 A0A151JB33 A0A151IJT3 A0A3L8D5E8 E2A3P4 A0A151WRL0 A0A158NKJ7 A0A195B101 A0A232FN17 A0A2J7PC65 K7IP26 A0A2P8XNE4 A0A0J7L0Z5 A0A164VVF7 A0A0P5AFK0 A0A336N175 W8C222 A0A336L593 A0A1B6IQN5 A0A182GHY3 A0A3R7MT98 A0A0P5D970 A0A0P6AL66 A0A1B6DIG1 A0A1A9WA67 A0A034VVZ5 A0A1Q3F1W9 A0A0A1WPY1 Q16M30 A0A1A9XKF7 A0A0L0C9R1 A0A1B0FHT8 A0A0K8VKF7 A0A0P6GVK2 A0A1A9VND4 A0A1B0AFD7 W5MWV5 A0A2C9LAH2 A0A0P7XLU3 A0A1W4ZYW9 A0A1B0AUD6 E9HKB2 A0A084W787 A0A3Q2PKK0 B3N3Z5 A0A1A8ERE7 A0A146P5K9 A0A1A8C4I8 A0A1A8NF67 A0A1A8V9B8 A0A1A8IM59 A0A1I8Q2W7 A0A1A8KZJ6 A0A0K8TNY7 Q6NWA8 B3MMY0 A0A1B6HVS2 A0A1A8SMW3 Q8IGU1 A0A3B0K0U2 A0A1A8QK59 Q8IPA1 Q8IP99 A0A1B0CLA3 A0A1A7YYB0 B4Q3C2 C1C3F6 C4XVG3 A0A3B5LGQ2 B4P1W2 A0A3B4BXU7 M4AV59 A0A3B5KA04 A0A1B6KN84 A0A182IL80
H9IVI4 I4DPA6 A0A194PJB7 A0A2W1BRI2 S4NR67 A0A0L7K3C9 A0A067QY89 A0A2J7PC59 E0VAQ9 A0A0L7RG24 A0A0M8ZRT2 A0A2A3E9Q6 A0A088A274 A0A310SNF1 A0A154NW62 A0A0C9RAW3 E2BA55 F4WWC6 A0A195FAZ0 A0A151JB33 A0A151IJT3 A0A3L8D5E8 E2A3P4 A0A151WRL0 A0A158NKJ7 A0A195B101 A0A232FN17 A0A2J7PC65 K7IP26 A0A2P8XNE4 A0A0J7L0Z5 A0A164VVF7 A0A0P5AFK0 A0A336N175 W8C222 A0A336L593 A0A1B6IQN5 A0A182GHY3 A0A3R7MT98 A0A0P5D970 A0A0P6AL66 A0A1B6DIG1 A0A1A9WA67 A0A034VVZ5 A0A1Q3F1W9 A0A0A1WPY1 Q16M30 A0A1A9XKF7 A0A0L0C9R1 A0A1B0FHT8 A0A0K8VKF7 A0A0P6GVK2 A0A1A9VND4 A0A1B0AFD7 W5MWV5 A0A2C9LAH2 A0A0P7XLU3 A0A1W4ZYW9 A0A1B0AUD6 E9HKB2 A0A084W787 A0A3Q2PKK0 B3N3Z5 A0A1A8ERE7 A0A146P5K9 A0A1A8C4I8 A0A1A8NF67 A0A1A8V9B8 A0A1A8IM59 A0A1I8Q2W7 A0A1A8KZJ6 A0A0K8TNY7 Q6NWA8 B3MMY0 A0A1B6HVS2 A0A1A8SMW3 Q8IGU1 A0A3B0K0U2 A0A1A8QK59 Q8IPA1 Q8IP99 A0A1B0CLA3 A0A1A7YYB0 B4Q3C2 C1C3F6 C4XVG3 A0A3B5LGQ2 B4P1W2 A0A3B4BXU7 M4AV59 A0A3B5KA04 A0A1B6KN84 A0A182IL80
Pubmed
26354079
22118469
19121390
22651552
28756777
23622113
+ More
26227816 24845553 20566863 20798317 21719571 30249741 21347285 28648823 20075255 29403074 24495485 26483478 25348373 25830018 17510324 26108605 15562597 21292972 24438588 17994087 26369729 15520368 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 23542700 21551351
26227816 24845553 20566863 20798317 21719571 30249741 21347285 28648823 20075255 29403074 24495485 26483478 25348373 25830018 17510324 26108605 15562597 21292972 24438588 17994087 26369729 15520368 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 23542700 21551351
EMBL
KQ459700
KPJ20406.1
RSAL01000316
RVE42603.1
GDQN01003607
JAT87447.1
+ More
NWSH01000518 PCG75900.1 ODYU01002500 SOQ40058.1 AGBW02011630 OWR46404.1 BABH01001167 AK403498 BAM19746.1 KQ459606 KPI91180.1 KZ149983 PZC75757.1 GAIX01014562 JAA77998.1 JTDY01014797 KOB51992.1 KK853205 KDR09857.1 NEVH01027067 PNF13916.1 DS235012 EEB10465.1 KQ414601 KOC69686.1 KQ435881 KOX69820.1 KZ288311 PBC28455.1 KQ759843 OAD62592.1 KQ434772 KZC03919.1 GBYB01010237 JAG80004.1 GL446664 EFN87464.1 GL888406 EGI61468.1 KQ981693 KYN37790.1 KQ979182 KYN22323.1 KQ977294 KYN03896.1 QOIP01000013 RLU15459.1 GL436457 EFN71936.1 KQ982805 KYQ50484.1 ADTU01018869 KQ976692 KYM77879.1 NNAY01000022 OXU31858.1 PNF13917.1 PYGN01001658 PSN33516.1 LBMM01001444 KMQ96306.1 LRGB01001361 KZS12698.1 GDIP01199536 JAJ23866.1 UFQT01004329 SSX35651.1 GAMC01000678 JAC05878.1 UFQS01001066 UFQT01001066 SSX08820.1 SSX28732.1 GECU01018477 JAS89229.1 JXUM01064723 KQ562312 KXJ76178.1 QCYY01000297 ROT85364.1 GDIP01161048 JAJ62354.1 GDIP01027761 JAM75954.1 GEDC01011807 JAS25491.1 GAKP01012927 JAC46025.1 GFDL01013492 JAV21553.1 GBXI01013410 JAD00882.1 CH477882 EAT35387.1 JRES01000725 KNC28945.1 CCAG010015264 GDHF01012955 GDHF01002233 JAI39359.1 JAI50081.1 GDIQ01027810 JAN66927.1 AHAT01000168 AHAT01000169 AHAT01000170 JARO02001231 KPP76082.1 JXJN01003606 GL732667 EFX67854.1 ATLV01021173 KE525314 KFB46081.1 CH954177 EDV58847.1 HAEB01002920 HAEC01013451 SBQ49447.1 GCES01147083 JAQ39239.1 HADZ01009820 HAEA01015536 SBP73761.1 HAEG01002576 SBR67531.1 HAEJ01015964 SBS56421.1 HAED01011961 HAEE01007519 SBQ98355.1 HAEF01000598 SBR37980.1 GDAI01002008 JAI15595.1 AY423005 BC067660 BC153454 CH902620 EDV31005.1 GECU01028917 JAS78789.1 HAEI01017242 SBS19711.1 BT001603 AAN71358.1 OUUW01000004 SPP79569.1 HAEH01011926 SBR93494.1 AE014134 BT015976 AAN10793.1 AAV36861.1 AAN10796.1 AHN54372.1 AJWK01017057 HADW01015525 HADX01012904 SBP35136.1 CM000361 CM002910 EDX04753.1 KMY89851.1 BT081385 ACO51515.1 BT088767 ACR55698.1 CM000157 EDW88133.1 GEBQ01027062 JAT12915.1
NWSH01000518 PCG75900.1 ODYU01002500 SOQ40058.1 AGBW02011630 OWR46404.1 BABH01001167 AK403498 BAM19746.1 KQ459606 KPI91180.1 KZ149983 PZC75757.1 GAIX01014562 JAA77998.1 JTDY01014797 KOB51992.1 KK853205 KDR09857.1 NEVH01027067 PNF13916.1 DS235012 EEB10465.1 KQ414601 KOC69686.1 KQ435881 KOX69820.1 KZ288311 PBC28455.1 KQ759843 OAD62592.1 KQ434772 KZC03919.1 GBYB01010237 JAG80004.1 GL446664 EFN87464.1 GL888406 EGI61468.1 KQ981693 KYN37790.1 KQ979182 KYN22323.1 KQ977294 KYN03896.1 QOIP01000013 RLU15459.1 GL436457 EFN71936.1 KQ982805 KYQ50484.1 ADTU01018869 KQ976692 KYM77879.1 NNAY01000022 OXU31858.1 PNF13917.1 PYGN01001658 PSN33516.1 LBMM01001444 KMQ96306.1 LRGB01001361 KZS12698.1 GDIP01199536 JAJ23866.1 UFQT01004329 SSX35651.1 GAMC01000678 JAC05878.1 UFQS01001066 UFQT01001066 SSX08820.1 SSX28732.1 GECU01018477 JAS89229.1 JXUM01064723 KQ562312 KXJ76178.1 QCYY01000297 ROT85364.1 GDIP01161048 JAJ62354.1 GDIP01027761 JAM75954.1 GEDC01011807 JAS25491.1 GAKP01012927 JAC46025.1 GFDL01013492 JAV21553.1 GBXI01013410 JAD00882.1 CH477882 EAT35387.1 JRES01000725 KNC28945.1 CCAG010015264 GDHF01012955 GDHF01002233 JAI39359.1 JAI50081.1 GDIQ01027810 JAN66927.1 AHAT01000168 AHAT01000169 AHAT01000170 JARO02001231 KPP76082.1 JXJN01003606 GL732667 EFX67854.1 ATLV01021173 KE525314 KFB46081.1 CH954177 EDV58847.1 HAEB01002920 HAEC01013451 SBQ49447.1 GCES01147083 JAQ39239.1 HADZ01009820 HAEA01015536 SBP73761.1 HAEG01002576 SBR67531.1 HAEJ01015964 SBS56421.1 HAED01011961 HAEE01007519 SBQ98355.1 HAEF01000598 SBR37980.1 GDAI01002008 JAI15595.1 AY423005 BC067660 BC153454 CH902620 EDV31005.1 GECU01028917 JAS78789.1 HAEI01017242 SBS19711.1 BT001603 AAN71358.1 OUUW01000004 SPP79569.1 HAEH01011926 SBR93494.1 AE014134 BT015976 AAN10793.1 AAV36861.1 AAN10796.1 AHN54372.1 AJWK01017057 HADW01015525 HADX01012904 SBP35136.1 CM000361 CM002910 EDX04753.1 KMY89851.1 BT081385 ACO51515.1 BT088767 ACR55698.1 CM000157 EDW88133.1 GEBQ01027062 JAT12915.1
Proteomes
UP000053240
UP000283053
UP000218220
UP000007151
UP000005204
UP000053268
+ More
UP000037510 UP000027135 UP000235965 UP000009046 UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000008237 UP000007755 UP000078541 UP000078492 UP000078542 UP000279307 UP000000311 UP000075809 UP000005205 UP000078540 UP000215335 UP000002358 UP000245037 UP000036403 UP000076858 UP000069940 UP000249989 UP000283509 UP000091820 UP000008820 UP000092443 UP000037069 UP000092444 UP000078200 UP000092445 UP000018468 UP000076420 UP000034805 UP000192224 UP000092460 UP000000305 UP000030765 UP000265000 UP000008711 UP000095300 UP000000437 UP000007801 UP000268350 UP000000803 UP000092461 UP000000304 UP000261380 UP000002282 UP000261440 UP000002852 UP000005226 UP000075880
UP000037510 UP000027135 UP000235965 UP000009046 UP000053825 UP000053105 UP000242457 UP000005203 UP000076502 UP000008237 UP000007755 UP000078541 UP000078492 UP000078542 UP000279307 UP000000311 UP000075809 UP000005205 UP000078540 UP000215335 UP000002358 UP000245037 UP000036403 UP000076858 UP000069940 UP000249989 UP000283509 UP000091820 UP000008820 UP000092443 UP000037069 UP000092444 UP000078200 UP000092445 UP000018468 UP000076420 UP000034805 UP000192224 UP000092460 UP000000305 UP000030765 UP000265000 UP000008711 UP000095300 UP000000437 UP000007801 UP000268350 UP000000803 UP000092461 UP000000304 UP000261380 UP000002282 UP000261440 UP000002852 UP000005226 UP000075880
Pfam
PF12767 SAGA-Tad1
SUPFAM
SSF47473
SSF47473
Gene 3D
ProteinModelPortal
A0A194RW40
A0A3S2TD59
A0A1E1WKL1
A0A2A4JWW4
A0A2H1VGV9
A0A212EY78
+ More
H9IVI4 I4DPA6 A0A194PJB7 A0A2W1BRI2 S4NR67 A0A0L7K3C9 A0A067QY89 A0A2J7PC59 E0VAQ9 A0A0L7RG24 A0A0M8ZRT2 A0A2A3E9Q6 A0A088A274 A0A310SNF1 A0A154NW62 A0A0C9RAW3 E2BA55 F4WWC6 A0A195FAZ0 A0A151JB33 A0A151IJT3 A0A3L8D5E8 E2A3P4 A0A151WRL0 A0A158NKJ7 A0A195B101 A0A232FN17 A0A2J7PC65 K7IP26 A0A2P8XNE4 A0A0J7L0Z5 A0A164VVF7 A0A0P5AFK0 A0A336N175 W8C222 A0A336L593 A0A1B6IQN5 A0A182GHY3 A0A3R7MT98 A0A0P5D970 A0A0P6AL66 A0A1B6DIG1 A0A1A9WA67 A0A034VVZ5 A0A1Q3F1W9 A0A0A1WPY1 Q16M30 A0A1A9XKF7 A0A0L0C9R1 A0A1B0FHT8 A0A0K8VKF7 A0A0P6GVK2 A0A1A9VND4 A0A1B0AFD7 W5MWV5 A0A2C9LAH2 A0A0P7XLU3 A0A1W4ZYW9 A0A1B0AUD6 E9HKB2 A0A084W787 A0A3Q2PKK0 B3N3Z5 A0A1A8ERE7 A0A146P5K9 A0A1A8C4I8 A0A1A8NF67 A0A1A8V9B8 A0A1A8IM59 A0A1I8Q2W7 A0A1A8KZJ6 A0A0K8TNY7 Q6NWA8 B3MMY0 A0A1B6HVS2 A0A1A8SMW3 Q8IGU1 A0A3B0K0U2 A0A1A8QK59 Q8IPA1 Q8IP99 A0A1B0CLA3 A0A1A7YYB0 B4Q3C2 C1C3F6 C4XVG3 A0A3B5LGQ2 B4P1W2 A0A3B4BXU7 M4AV59 A0A3B5KA04 A0A1B6KN84 A0A182IL80
H9IVI4 I4DPA6 A0A194PJB7 A0A2W1BRI2 S4NR67 A0A0L7K3C9 A0A067QY89 A0A2J7PC59 E0VAQ9 A0A0L7RG24 A0A0M8ZRT2 A0A2A3E9Q6 A0A088A274 A0A310SNF1 A0A154NW62 A0A0C9RAW3 E2BA55 F4WWC6 A0A195FAZ0 A0A151JB33 A0A151IJT3 A0A3L8D5E8 E2A3P4 A0A151WRL0 A0A158NKJ7 A0A195B101 A0A232FN17 A0A2J7PC65 K7IP26 A0A2P8XNE4 A0A0J7L0Z5 A0A164VVF7 A0A0P5AFK0 A0A336N175 W8C222 A0A336L593 A0A1B6IQN5 A0A182GHY3 A0A3R7MT98 A0A0P5D970 A0A0P6AL66 A0A1B6DIG1 A0A1A9WA67 A0A034VVZ5 A0A1Q3F1W9 A0A0A1WPY1 Q16M30 A0A1A9XKF7 A0A0L0C9R1 A0A1B0FHT8 A0A0K8VKF7 A0A0P6GVK2 A0A1A9VND4 A0A1B0AFD7 W5MWV5 A0A2C9LAH2 A0A0P7XLU3 A0A1W4ZYW9 A0A1B0AUD6 E9HKB2 A0A084W787 A0A3Q2PKK0 B3N3Z5 A0A1A8ERE7 A0A146P5K9 A0A1A8C4I8 A0A1A8NF67 A0A1A8V9B8 A0A1A8IM59 A0A1I8Q2W7 A0A1A8KZJ6 A0A0K8TNY7 Q6NWA8 B3MMY0 A0A1B6HVS2 A0A1A8SMW3 Q8IGU1 A0A3B0K0U2 A0A1A8QK59 Q8IPA1 Q8IP99 A0A1B0CLA3 A0A1A7YYB0 B4Q3C2 C1C3F6 C4XVG3 A0A3B5LGQ2 B4P1W2 A0A3B4BXU7 M4AV59 A0A3B5KA04 A0A1B6KN84 A0A182IL80
Ontologies
GO
PANTHER
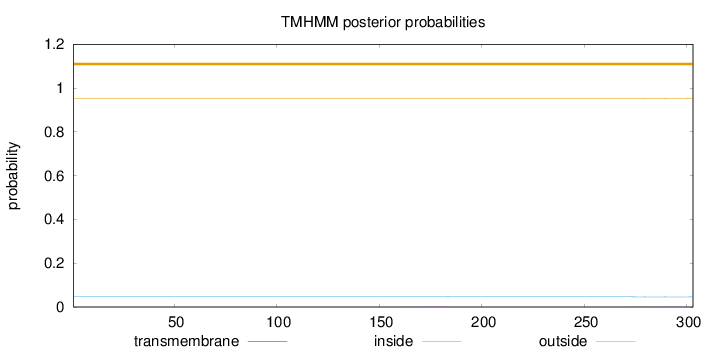
Topology
Subcellular location
Nucleus
Length:
303
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03361
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.04642
outside
1 - 303
Population Genetic Test Statistics
Pi
313.442459
Theta
189.369067
Tajima's D
1.692589
CLR
0
CSRT
0.830508474576271
Interpretation
Uncertain
