Gene
KWMTBOMO07724
Pre Gene Modal
BGIBMGA000880
Annotation
PREDICTED:_ATP-dependent_RNA_helicase_SUV3_homolog?_mitochondrial_[Bombyx_mori]
Full name
ATP-dependent RNA helicase SUV3 homolog, mitochondrial
Location in the cell
Cytoplasmic Reliability : 1.478 Nuclear Reliability : 2.068
Sequence
CDS
ATGTATAAAGGGTTTTCCAGAATATTAGCCCTTCATAATGTAGAAAAATTACAACTTAATGTATCAAATCTCAGATGTAATATCTTGTGCGCCCGTAAACTCAAACCCGCCGGTATAGGTTATATTTTGGTAAGAACTAAAAAACATGATCGGAATCCTTCTGCTTTATTTGTGCCAGTGCCAGTGAAACCAAACTCCGATGACATAAACATTGGTGAAGAATTTACAGGACGTCTAAAAAAACAAGACTTGTTAAAAATTCTTAACAAATTTTACCAAAAACCTGAAATCAGAGTGCTTGCTTCAGAAAATGGATTGGATGATCAATTGTTACATCAAGCTTTCCTTTCATTCAGACGGCATTGTTTAGAACATGATTTACCACCAGACTTACATATTACGATCAGTGATATACTTCAAGGTGCAGGACATGTTGATGATTTGTTTCCATACTTCTTGAGGCATGCAAGACGAGCATTTCCGCACCTTGACTGCTTGGAAGACTTGAAAAAGATATCAGATTTGAGAACACCAGCAAACTGGTATCCTGAAGCTCGAAGTATGGATCGCAAAATTATATTTCATGCAGGTCCCACAAATTCAGGAAAAACTTATCATGCAATGGAGAGATTCTTAAAATCGAAATCTGGTGTGTACTGTGGTCCCTTAAAGTTACTAGCAACTGAGATATATCATAAATCGAATAAAAGTGGCACACCCTGCGATCTAGTAACAGGAGAAGAAAGAAGGCACGCTTCATTATACAACACCTTAATAAATGGTACAGATGATAATGACATAGAAACATCAGATGTCGAACCTTATTTGTTAGAAGAAACCGAATTAACACCGTCTGGTCATGTTGCATGTACAGTTGAAATGACTTCTCTAAATAATAAATATGAAGTGGCAATCATAGATGAAATACAAATGATAGGAGATAGAGGAAGAGGGTGGGCATGGACTCGAGCTATATTAGGTTTACAAGCAGATGAAATACATTTATGTGGGGAGGCAGGTGCAATAAATCTCATTGAAGAAATTTGTAACACAACCGGTGAAGTAATGGAGGTTCGTTCGTACAAAAGATTAACACAGCTAAAAGTCGAAGATACCGCTTTAGGATCATTAGACAATGTTCAGCCCGGAGACTGTATTGTGTGTTTCAATAAAAACGACATTTACAGTGTAAGTCGAGCCATTGAACAGAGAGGACATGAGGTAGCAGTTATCTATGGCAGTTTACCACCTGGTACTAAGCTAGCTCAGGCAAACAAGTTCAACGATCCGGAGAGTTCGTGTAAAGTTATGGTTGCGACAGACGCCATTGGACTCGGTATAAATTTGAGCATCCGGCGGATAATCTTTTACTCATTAATCAAACCGGTAATTAATGAAGACGGTGAGAAAGAGATGGACGTTATTAGTATATCACAAGCGTTGCAGATTGCAGGTCGGGCCGGTCGTTACGGCAGCGCGTGGGAGACCGGCCACGTGACCTCGTACCGGCCCGAAGACCTCGCGACGCTGAAGACGCTGCTGTCCCAGCCGCCCGAGCCGGTCACGCAGGCCGGCCTGCACCCCACCTCCGAGCAGATGGAGCTGTACGCCTACCATCTGCCCCACGCCACTCTCAGCAGTCTCATGGACATATTCGTTCACTTGTGCACGGTCGACGATTCGCTTTACTTCATGTGCAACACGGAGGGGTTCAAGTTCTTAGCGGAGATGATACAGCACGTTCCGTTGCCGCTGCGGGCGCGCTACGTGTTCTGTTGCGCGCCCATCAATAACAAGCTACCGTTCGTTTGTGCTACCTTCCTGAAAATGGTGAGACAGTACAGTCGCAACGAGCCCATCACCCGGAACTGGCTGAGCGGCACAGTGGAGTGGCCGCTGCCCTCTCCTAGGACCATACTGGACCTCGTGCACTTGGAATCTGTCTTTGACGTCCTAGAGCTTTACTTGTGGTTAAGCTATCGATTTCCGGACATGTTTCCGGACGTGAAATTAGTTCGTGACATGGAGACTGAATTAGATGCTATCATTCAACAAGGAATTTTCCAAATAACTAGGTTACTAAGAAATTCTGAACAGATGATCCGCGATGAGGACAGCGGCTTTGCGATCGGCCACGGCAGCAAGAGGGTCAATAAGATGCTGGCCGGACAATCAATGGGCGAGGAGAAGGGAAAATTGTCGGAATTACTAGTTGCCAGGGGACTCATAACGCCGCAGATGCTGAAGAAACTTCAGCAAGAACTCTCCACGGACAAGAAGATAGACAGAACGAAAAAGAATAGGAACAGAAGGAAATAA
Protein
MYKGFSRILALHNVEKLQLNVSNLRCNILCARKLKPAGIGYILVRTKKHDRNPSALFVPVPVKPNSDDINIGEEFTGRLKKQDLLKILNKFYQKPEIRVLASENGLDDQLLHQAFLSFRRHCLEHDLPPDLHITISDILQGAGHVDDLFPYFLRHARRAFPHLDCLEDLKKISDLRTPANWYPEARSMDRKIIFHAGPTNSGKTYHAMERFLKSKSGVYCGPLKLLATEIYHKSNKSGTPCDLVTGEERRHASLYNTLINGTDDNDIETSDVEPYLLEETELTPSGHVACTVEMTSLNNKYEVAIIDEIQMIGDRGRGWAWTRAILGLQADEIHLCGEAGAINLIEEICNTTGEVMEVRSYKRLTQLKVEDTALGSLDNVQPGDCIVCFNKNDIYSVSRAIEQRGHEVAVIYGSLPPGTKLAQANKFNDPESSCKVMVATDAIGLGINLSIRRIIFYSLIKPVINEDGEKEMDVISISQALQIAGRAGRYGSAWETGHVTSYRPEDLATLKTLLSQPPEPVTQAGLHPTSEQMELYAYHLPHATLSSLMDIFVHLCTVDDSLYFMCNTEGFKFLAEMIQHVPLPLRARYVFCCAPINNKLPFVCATFLKMVRQYSRNEPITRNWLSGTVEWPLPSPRTILDLVHLESVFDVLELYLWLSYRFPDMFPDVKLVRDMETELDAIIQQGIFQITRLLRNSEQMIRDEDSGFAIGHGSKRVNKMLAGQSMGEEKGKLSELLVARGLITPQMLKKLQQELSTDKKIDRTKKNRNRRK
Summary
Description
Major helicase player in mitochondrial RNA metabolism and maintenance (PubMed:26152302). Likely component of the mitochondrial degradosome (mtEXO) complex, that degrades 3' overhang double-stranded RNA with a 3'-to-5' directionality in an ATP-dependent manner (By similarity). ATPase and ATP-dependent multisubstrate helicase, able to unwind double-stranded (ds) DNA and RNA, and RNA/DNA heteroduplexes in the 5'-to-3' direction (By similarity). Regulates mRNA stability and is required for the correct processing and maturation of mitochondrial transcripts (PubMed:26152302).
Major helicase player in mitochondrial RNA metabolism and maintenance. Likely component of the mitochondrial degradosome (mtEXO) complex, that degrades 3' overhang double-stranded RNA with a 3'-to-5' directionality in an ATP-dependent manner. ATPase and ATP-dependent multisubstrate helicase, able to unwind double-stranded (ds) DNA and RNA, and RNA/DNA heteroduplexes in the 5'-to-3' direction. Regulates mRNA stability and is required for the correct processing and maturation of mitochondrial transcripts.
Major helicase player in mitochondrial RNA metabolism and maintenance. Likely component of the mitochondrial degradosome (mtEXO) complex, that degrades 3' overhang double-stranded RNA with a 3'-to-5' directionality in an ATP-dependent manner. ATPase and ATP-dependent multisubstrate helicase, able to unwind double-stranded (ds) DNA and RNA, and RNA/DNA heteroduplexes in the 5'-to-3' direction. Regulates mRNA stability and is required for the correct processing and maturation of mitochondrial transcripts.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Similarity
Belongs to the helicase family.
Keywords
ATP-binding
Complete proteome
Helicase
Hydrolase
Mitochondrion
Nucleotide-binding
Reference proteome
Transit peptide
Feature
chain ATP-dependent RNA helicase SUV3 homolog, mitochondrial
Uniprot
A0A2H1VGU2
A0A194PDT4
A0A212EY30
A0A2W1BL46
A0A194RRA1
A0A2A4J6E3
+ More
H9IUF0 A0A067QM73 A0A2J7PJG0 A0A1Y1MNL2 D6X1T4 U4UKE3 N6U3Z9 A0A336KF45 U4UTL8 A0A1W4W8F5 A0A2J7PJG6 B4KAA8 A0A182FL06 A0A182NSJ1 U4UQC8 A0A232F788 A0A2M4AH83 A0A0M4ERV6 A0A1B6MUP0 A0A084VDU1 W8BCU4 A0A182JHC5 W5J3X1 A0A0Q9WEH1 B4M699 A0A1B6D0T0 A0A1S4FGQ2 Q171V5 A0A182R893 B0WAJ5 A0A0L0CLV1 A0A182YLL8 A0A182KFR9 A0A182LSQ6 A0A182P089 A0A182VS47 A0A182GF47 A0A182QWD7 A0A182SDR8 A0A2H8TJL1 A0A1Q3FEV0 A0A1W4VLT5 A0A1I8PCV7 A0A182UUD3 J9K0T4 A0A2S2PHI2 A0A182WVZ9 A0A1B0FFY6 B4N923 A0A182TZU8 B4JI86 A0A1A9ZT33 B4PTQ6 Q7QH43 A0A1S4GIX1 Q9VN03 A0A3B0KIG4 A0A1A9UFI9 A0A1B6EMZ4 Q295E6 B4GM55 B4QV00 A0A1A9WC08 B3P1Y8 A0A1A9YQ02 A0A1B0B4Z9 A0A195BRU5 A0A1J1J5U4 A0A158NZW4 A0A1Z5KXW9 A0A3L8DQA6 E2A4R8 A0A195F3J1 E9I9I9 E2B4B5 A0A026WC11 A0A293MTP4 A0A0P6DME9 A0A0P5QQT2 A0A0N8DYS8 A0A2R5LJF9 A0A151XEP4 A0A2P6KX59 A0A0P5G6A7 A0A0P5EQP9 A0A0P5U9R3 A0A195EDL9 A0A0P5GMV8 E9HJW5 A0A0P6HDC7 A0A023GLU5 G3MRG0 V5I2W0 B7PKK6
H9IUF0 A0A067QM73 A0A2J7PJG0 A0A1Y1MNL2 D6X1T4 U4UKE3 N6U3Z9 A0A336KF45 U4UTL8 A0A1W4W8F5 A0A2J7PJG6 B4KAA8 A0A182FL06 A0A182NSJ1 U4UQC8 A0A232F788 A0A2M4AH83 A0A0M4ERV6 A0A1B6MUP0 A0A084VDU1 W8BCU4 A0A182JHC5 W5J3X1 A0A0Q9WEH1 B4M699 A0A1B6D0T0 A0A1S4FGQ2 Q171V5 A0A182R893 B0WAJ5 A0A0L0CLV1 A0A182YLL8 A0A182KFR9 A0A182LSQ6 A0A182P089 A0A182VS47 A0A182GF47 A0A182QWD7 A0A182SDR8 A0A2H8TJL1 A0A1Q3FEV0 A0A1W4VLT5 A0A1I8PCV7 A0A182UUD3 J9K0T4 A0A2S2PHI2 A0A182WVZ9 A0A1B0FFY6 B4N923 A0A182TZU8 B4JI86 A0A1A9ZT33 B4PTQ6 Q7QH43 A0A1S4GIX1 Q9VN03 A0A3B0KIG4 A0A1A9UFI9 A0A1B6EMZ4 Q295E6 B4GM55 B4QV00 A0A1A9WC08 B3P1Y8 A0A1A9YQ02 A0A1B0B4Z9 A0A195BRU5 A0A1J1J5U4 A0A158NZW4 A0A1Z5KXW9 A0A3L8DQA6 E2A4R8 A0A195F3J1 E9I9I9 E2B4B5 A0A026WC11 A0A293MTP4 A0A0P6DME9 A0A0P5QQT2 A0A0N8DYS8 A0A2R5LJF9 A0A151XEP4 A0A2P6KX59 A0A0P5G6A7 A0A0P5EQP9 A0A0P5U9R3 A0A195EDL9 A0A0P5GMV8 E9HJW5 A0A0P6HDC7 A0A023GLU5 G3MRG0 V5I2W0 B7PKK6
EC Number
3.6.4.13
Pubmed
26354079
22118469
28756777
19121390
24845553
28004739
+ More
18362917 19820115 23537049 17994087 28648823 24438588 24495485 20920257 23761445 17510324 26108605 25244985 26483478 17550304 12364791 10731132 12537572 12537569 26152302 15632085 21347285 28528879 30249741 20798317 21282665 24508170 21292972 22216098 25765539
18362917 19820115 23537049 17994087 28648823 24438588 24495485 20920257 23761445 17510324 26108605 25244985 26483478 17550304 12364791 10731132 12537572 12537569 26152302 15632085 21347285 28528879 30249741 20798317 21282665 24508170 21292972 22216098 25765539
EMBL
ODYU01002500
SOQ40060.1
KQ459606
KPI91178.1
AGBW02011630
OWR46402.1
+ More
KZ149983 PZC75759.1 KQ459700 KPJ20408.1 NWSH01003023 PCG67094.1 BABH01001167 KK853179 KDR10211.1 NEVH01024949 PNF16463.1 GEZM01026090 JAV87252.1 KQ971371 EFA10159.1 KB630054 KB632237 ERL83363.1 ERL90465.1 APGK01043589 KB741015 ENN75346.1 UFQS01000350 UFQT01000350 SSX03081.1 SSX23447.1 KI208369 ERL95848.1 PNF16458.1 CH933806 EDW14595.2 KI210459 ERL96274.1 NNAY01000743 OXU26724.1 GGFK01006834 MBW40155.1 CP012526 ALC45255.1 GEBQ01000339 JAT39638.1 ATLV01011796 KE524723 KFB36135.1 GAMC01007460 JAB99095.1 ADMH02002130 ETN58541.1 CH940652 KRF78736.1 EDW59175.1 GEDC01028125 GEDC01018024 JAS09173.1 JAS19274.1 CH477445 EAT40781.1 DS231872 EDS41418.1 JRES01000310 KNC32404.1 AXCM01002011 JXUM01059133 KQ562037 KXJ76843.1 AXCN02000234 GFXV01002522 MBW14327.1 GFDL01008968 JAV26077.1 ABLF02038337 GGMR01016258 MBY28877.1 CCAG010004727 CH964232 EDW81570.1 CH916369 EDV92967.1 CM000160 EDW95639.2 AAAB01008820 EAA05399.5 AE014297 AY058527 BT133067 AEX93152.1 OUUW01000014 SPP88300.1 GECZ01030465 JAS39304.1 CM000070 CH479185 EDW37929.1 CM000364 EDX11581.1 CH954181 EDV47761.1 JXJN01008576 KQ976423 KYM88740.1 CVRI01000070 CRL07148.1 ADTU01000904 ADTU01000905 GFJQ02007031 JAV99938.1 QOIP01000005 RLU22620.1 GL436716 EFN71602.1 KQ981855 KYN34951.1 GL761812 EFZ22765.1 GL445549 EFN89463.1 KK107293 EZA53186.1 GFWV01018580 MAA43308.1 GDIQ01076142 JAN18595.1 GDIQ01110840 JAL40886.1 GDIQ01078644 LRGB01000005 JAN16093.1 KZS22089.1 GGLE01005556 MBY09682.1 KQ982231 KYQ58864.1 MWRG01004014 PRD30914.1 GDIQ01253232 JAJ98492.1 GDIP01143172 JAJ80230.1 GDIP01133330 JAL70384.1 KQ979039 KYN23320.1 GDIQ01244998 JAK06727.1 GL732665 EFX67971.1 GDIQ01021130 JAN73607.1 GBBM01000609 JAC34809.1 JO844461 AEO36078.1 GANP01000526 JAB83942.1 ABJB011085262 DS734517 EEC07128.1
KZ149983 PZC75759.1 KQ459700 KPJ20408.1 NWSH01003023 PCG67094.1 BABH01001167 KK853179 KDR10211.1 NEVH01024949 PNF16463.1 GEZM01026090 JAV87252.1 KQ971371 EFA10159.1 KB630054 KB632237 ERL83363.1 ERL90465.1 APGK01043589 KB741015 ENN75346.1 UFQS01000350 UFQT01000350 SSX03081.1 SSX23447.1 KI208369 ERL95848.1 PNF16458.1 CH933806 EDW14595.2 KI210459 ERL96274.1 NNAY01000743 OXU26724.1 GGFK01006834 MBW40155.1 CP012526 ALC45255.1 GEBQ01000339 JAT39638.1 ATLV01011796 KE524723 KFB36135.1 GAMC01007460 JAB99095.1 ADMH02002130 ETN58541.1 CH940652 KRF78736.1 EDW59175.1 GEDC01028125 GEDC01018024 JAS09173.1 JAS19274.1 CH477445 EAT40781.1 DS231872 EDS41418.1 JRES01000310 KNC32404.1 AXCM01002011 JXUM01059133 KQ562037 KXJ76843.1 AXCN02000234 GFXV01002522 MBW14327.1 GFDL01008968 JAV26077.1 ABLF02038337 GGMR01016258 MBY28877.1 CCAG010004727 CH964232 EDW81570.1 CH916369 EDV92967.1 CM000160 EDW95639.2 AAAB01008820 EAA05399.5 AE014297 AY058527 BT133067 AEX93152.1 OUUW01000014 SPP88300.1 GECZ01030465 JAS39304.1 CM000070 CH479185 EDW37929.1 CM000364 EDX11581.1 CH954181 EDV47761.1 JXJN01008576 KQ976423 KYM88740.1 CVRI01000070 CRL07148.1 ADTU01000904 ADTU01000905 GFJQ02007031 JAV99938.1 QOIP01000005 RLU22620.1 GL436716 EFN71602.1 KQ981855 KYN34951.1 GL761812 EFZ22765.1 GL445549 EFN89463.1 KK107293 EZA53186.1 GFWV01018580 MAA43308.1 GDIQ01076142 JAN18595.1 GDIQ01110840 JAL40886.1 GDIQ01078644 LRGB01000005 JAN16093.1 KZS22089.1 GGLE01005556 MBY09682.1 KQ982231 KYQ58864.1 MWRG01004014 PRD30914.1 GDIQ01253232 JAJ98492.1 GDIP01143172 JAJ80230.1 GDIP01133330 JAL70384.1 KQ979039 KYN23320.1 GDIQ01244998 JAK06727.1 GL732665 EFX67971.1 GDIQ01021130 JAN73607.1 GBBM01000609 JAC34809.1 JO844461 AEO36078.1 GANP01000526 JAB83942.1 ABJB011085262 DS734517 EEC07128.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000005204
UP000027135
+ More
UP000235965 UP000007266 UP000030742 UP000019118 UP000192223 UP000009192 UP000069272 UP000075884 UP000215335 UP000092553 UP000030765 UP000075880 UP000000673 UP000008792 UP000008820 UP000075900 UP000002320 UP000037069 UP000076408 UP000075881 UP000075883 UP000075885 UP000075920 UP000069940 UP000249989 UP000075886 UP000075901 UP000192221 UP000095300 UP000075903 UP000007819 UP000076407 UP000092444 UP000007798 UP000075902 UP000001070 UP000092445 UP000002282 UP000007062 UP000000803 UP000268350 UP000078200 UP000001819 UP000008744 UP000000304 UP000091820 UP000008711 UP000092443 UP000092460 UP000078540 UP000183832 UP000005205 UP000279307 UP000000311 UP000078541 UP000008237 UP000053097 UP000076858 UP000075809 UP000078492 UP000000305 UP000001555
UP000235965 UP000007266 UP000030742 UP000019118 UP000192223 UP000009192 UP000069272 UP000075884 UP000215335 UP000092553 UP000030765 UP000075880 UP000000673 UP000008792 UP000008820 UP000075900 UP000002320 UP000037069 UP000076408 UP000075881 UP000075883 UP000075885 UP000075920 UP000069940 UP000249989 UP000075886 UP000075901 UP000192221 UP000095300 UP000075903 UP000007819 UP000076407 UP000092444 UP000007798 UP000075902 UP000001070 UP000092445 UP000002282 UP000007062 UP000000803 UP000268350 UP000078200 UP000001819 UP000008744 UP000000304 UP000091820 UP000008711 UP000092443 UP000092460 UP000078540 UP000183832 UP000005205 UP000279307 UP000000311 UP000078541 UP000008237 UP000053097 UP000076858 UP000075809 UP000078492 UP000000305 UP000001555
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2H1VGU2
A0A194PDT4
A0A212EY30
A0A2W1BL46
A0A194RRA1
A0A2A4J6E3
+ More
H9IUF0 A0A067QM73 A0A2J7PJG0 A0A1Y1MNL2 D6X1T4 U4UKE3 N6U3Z9 A0A336KF45 U4UTL8 A0A1W4W8F5 A0A2J7PJG6 B4KAA8 A0A182FL06 A0A182NSJ1 U4UQC8 A0A232F788 A0A2M4AH83 A0A0M4ERV6 A0A1B6MUP0 A0A084VDU1 W8BCU4 A0A182JHC5 W5J3X1 A0A0Q9WEH1 B4M699 A0A1B6D0T0 A0A1S4FGQ2 Q171V5 A0A182R893 B0WAJ5 A0A0L0CLV1 A0A182YLL8 A0A182KFR9 A0A182LSQ6 A0A182P089 A0A182VS47 A0A182GF47 A0A182QWD7 A0A182SDR8 A0A2H8TJL1 A0A1Q3FEV0 A0A1W4VLT5 A0A1I8PCV7 A0A182UUD3 J9K0T4 A0A2S2PHI2 A0A182WVZ9 A0A1B0FFY6 B4N923 A0A182TZU8 B4JI86 A0A1A9ZT33 B4PTQ6 Q7QH43 A0A1S4GIX1 Q9VN03 A0A3B0KIG4 A0A1A9UFI9 A0A1B6EMZ4 Q295E6 B4GM55 B4QV00 A0A1A9WC08 B3P1Y8 A0A1A9YQ02 A0A1B0B4Z9 A0A195BRU5 A0A1J1J5U4 A0A158NZW4 A0A1Z5KXW9 A0A3L8DQA6 E2A4R8 A0A195F3J1 E9I9I9 E2B4B5 A0A026WC11 A0A293MTP4 A0A0P6DME9 A0A0P5QQT2 A0A0N8DYS8 A0A2R5LJF9 A0A151XEP4 A0A2P6KX59 A0A0P5G6A7 A0A0P5EQP9 A0A0P5U9R3 A0A195EDL9 A0A0P5GMV8 E9HJW5 A0A0P6HDC7 A0A023GLU5 G3MRG0 V5I2W0 B7PKK6
H9IUF0 A0A067QM73 A0A2J7PJG0 A0A1Y1MNL2 D6X1T4 U4UKE3 N6U3Z9 A0A336KF45 U4UTL8 A0A1W4W8F5 A0A2J7PJG6 B4KAA8 A0A182FL06 A0A182NSJ1 U4UQC8 A0A232F788 A0A2M4AH83 A0A0M4ERV6 A0A1B6MUP0 A0A084VDU1 W8BCU4 A0A182JHC5 W5J3X1 A0A0Q9WEH1 B4M699 A0A1B6D0T0 A0A1S4FGQ2 Q171V5 A0A182R893 B0WAJ5 A0A0L0CLV1 A0A182YLL8 A0A182KFR9 A0A182LSQ6 A0A182P089 A0A182VS47 A0A182GF47 A0A182QWD7 A0A182SDR8 A0A2H8TJL1 A0A1Q3FEV0 A0A1W4VLT5 A0A1I8PCV7 A0A182UUD3 J9K0T4 A0A2S2PHI2 A0A182WVZ9 A0A1B0FFY6 B4N923 A0A182TZU8 B4JI86 A0A1A9ZT33 B4PTQ6 Q7QH43 A0A1S4GIX1 Q9VN03 A0A3B0KIG4 A0A1A9UFI9 A0A1B6EMZ4 Q295E6 B4GM55 B4QV00 A0A1A9WC08 B3P1Y8 A0A1A9YQ02 A0A1B0B4Z9 A0A195BRU5 A0A1J1J5U4 A0A158NZW4 A0A1Z5KXW9 A0A3L8DQA6 E2A4R8 A0A195F3J1 E9I9I9 E2B4B5 A0A026WC11 A0A293MTP4 A0A0P6DME9 A0A0P5QQT2 A0A0N8DYS8 A0A2R5LJF9 A0A151XEP4 A0A2P6KX59 A0A0P5G6A7 A0A0P5EQP9 A0A0P5U9R3 A0A195EDL9 A0A0P5GMV8 E9HJW5 A0A0P6HDC7 A0A023GLU5 G3MRG0 V5I2W0 B7PKK6
PDB
3RC8
E-value=0,
Score=2125
Ontologies
GO
GO:0016817
GO:0004386
GO:0004004
GO:0000965
GO:0045025
GO:0006401
GO:0016021
GO:0005739
GO:0070131
GO:0022904
GO:0090646
GO:0006119
GO:0044528
GO:0097222
GO:0005524
GO:0003678
GO:0000963
GO:0032508
GO:0003677
GO:0005759
GO:0003924
GO:0016020
GO:0006508
GO:0016301
GO:0016573
GO:0008270
GO:0022891
GO:0003707
GO:0051537
Topology
Subcellular location
Mitochondrion
Unlike in mammals, does not localize to the nucleus. With evidence from 5 publications.
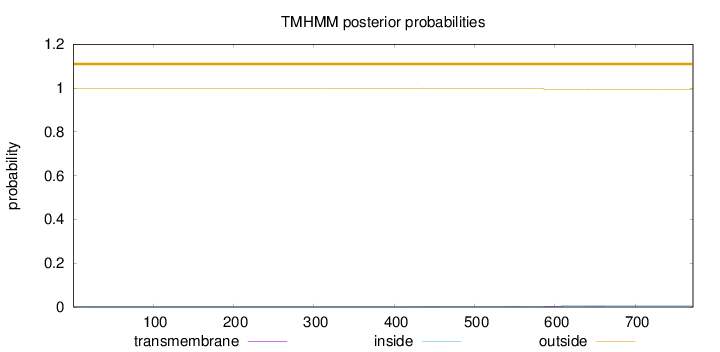
Length:
772
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15001
Exp number, first 60 AAs:
0.00079
Total prob of N-in:
0.00060
outside
1 - 772
Population Genetic Test Statistics
Pi
197.647878
Theta
151.733461
Tajima's D
0.973468
CLR
0.464288
CSRT
0.657417129143543
Interpretation
Uncertain
