Gene
KWMTBOMO07720
Pre Gene Modal
BGIBMGA000879
Annotation
PREDICTED:_retinal_rod_rhodopsin-sensitive_cGMP_3'?5'-cyclic_phosphodiesterase_subunit_delta_[Papilio_machaon]
Full name
Probable cGMP 3',5'-cyclic phosphodiesterase subunit delta
+ More
Phosphodiesterase delta-like protein
Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta
Phosphodiesterase delta-like protein
Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta
Alternative Name
Protein p17
Location in the cell
Nuclear Reliability : 2.199
Sequence
CDS
ATGACTGACGTTCTAACAACTGAACGAGATAAAAATGATCGTGTGGAGACTATTCTGAAGGGATTTCAAATTAATTGGATGAATCTTCGTGATGCTGACACAGGAAAAATTCTTTGGCAACATAATGAGGACATGTCAAGTCCAGATGCGGAACACGAAGCAAGAGTACCCAAAAGAATTCTCAAGTGCAGAGTAGTGTCTCGTGAAATGAACTTCAGCTCAATAGAATCCATGGATAGATTTAGATTAGAACAGAAAGTACTCTTCAAAGGGAGATGTTTAGAAGAGTGGTTTTTCGATTTTGGTTATGTGATCCCGAACTCAACGAACACTTGGCAATCCGTTATAGAGTCAGCGCCTGAATCGCAAATGATGCCGGCCAATGTGCTCAATGGGAACGTGGTTATCGAAACGAAATTTTTTGATGGAGATTTGCTGATCACAACCTCTAGAGTACGACTGTTCTATATATGA
Protein
MTDVLTTERDKNDRVETILKGFQINWMNLRDADTGKILWQHNEDMSSPDAEHEARVPKRILKCRVVSREMNFSSIESMDRFRLEQKVLFKGRCLEEWFFDFGYVIPNSTNTWQSVIESAPESQMMPANVLNGNVVIETKFFDGDLLITTSRVRLFYI
Summary
Description
Promotes the release of prenylated target proteins from cellular membranes. Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location. Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E. Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate.
Promotes the release of prenylated target proteins from cellular membranes (By similarity). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (By similarity). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (By similarity). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Promotes the release of prenylated target proteins from cellular membranes (PubMed:8798640). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (By similarity). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (By similarity). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Promotes the release of prenylated target proteins from cellular membranes (PubMed:9712853). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (PubMed:22002721, PubMed:23698361). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (PubMed:24166846). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Promotes the release of prenylated target proteins from cellular membranes (By similarity). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (By similarity). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (By similarity). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Promotes the release of prenylated target proteins from cellular membranes (PubMed:8798640). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (By similarity). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (By similarity). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Promotes the release of prenylated target proteins from cellular membranes (PubMed:9712853). Modulates the activity of prenylated or palmitoylated Ras family members by regulating their subcellular location (PubMed:22002721, PubMed:23698361). Required for normal ciliary targeting of farnesylated target proteins, such as INPP5E (PubMed:24166846). Modulates the subcellular location of target proteins by acting as a GTP specific dissociation inhibitor (GDI) (By similarity). Increases the affinity of ARL3 for GTP by several orders of magnitude. Stabilizes ARL3-GTP by decreasing the nucleotide dissociation rate (By similarity).
Subunit
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains and two inhibitory chains; has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane.
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (By similarity). Interacts with prenylated GRK1 and GRK7 (By similarity). Interacts with prenylated INPP5E (By similarity). Interacts with prenylated Ras family members, including HRAS, KRAS, NRAS, RAP2A, RAP2C and RHEB (By similarity). Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (By similarity). Interacts with RPGR (By similarity). Interacts with ARL2 (By similarity). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (By similarity). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (By similarity).
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (PubMed:8798640). Interacts with prenylated GRK1 and GRK7 (PubMed:14561760). Interacts with prenylated INPP5E (By similarity). Interacts with prenylated Ras family members, including HRAS, KRAS, NRAS, RAP2A, RAP2C and RHEB (By similarity). Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (By similarity). Interacts with RPGR (By similarity). Interacts with ARL2 (PubMed:14561760). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (By similarity). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (By similarity).
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (By similarity). Interacts with prenylated GRK1 and GRK7 (By similarity). Interacts with prenylated Ras family members, including RAP2A and RAP2C (By similarity). Interacts with prenylated RHEB and NRAS (PubMed:22002721). Interacts with prenylated HRAS and KRAS. Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (PubMed:9712853). Interacts with prenylated INPP5E (PubMed:24166846). Interacts with RPGR (PubMed:9990021, PubMed:24166846, PubMed:23559067). Interacts with ARL2 (PubMed:24166846, PubMed:22002721). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (PubMed:24166846). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (PubMed:22002721).
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (By similarity). Interacts with prenylated GRK1 and GRK7 (By similarity). Interacts with prenylated INPP5E (By similarity). Interacts with prenylated Ras family members, including HRAS, KRAS, NRAS, RAP2A, RAP2C and RHEB (By similarity). Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (By similarity). Interacts with RPGR (By similarity). Interacts with ARL2 (By similarity). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (By similarity). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (By similarity).
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (PubMed:8798640). Interacts with prenylated GRK1 and GRK7 (PubMed:14561760). Interacts with prenylated INPP5E (By similarity). Interacts with prenylated Ras family members, including HRAS, KRAS, NRAS, RAP2A, RAP2C and RHEB (By similarity). Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (By similarity). Interacts with RPGR (By similarity). Interacts with ARL2 (PubMed:14561760). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (By similarity). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (By similarity).
Interacts with the prenylated catalytic subunits of PDE6, an oligomer composed of two catalytic chains (PDE6A and PDE6B) and two inhibitory chains (gamma); has no effect on enzyme activity but promotes the release of the prenylated enzyme from cell membrane (By similarity). Interacts with prenylated GRK1 and GRK7 (By similarity). Interacts with prenylated Ras family members, including RAP2A and RAP2C (By similarity). Interacts with prenylated RHEB and NRAS (PubMed:22002721). Interacts with prenylated HRAS and KRAS. Interacts with RAB13 (prenylated form); dissociates RAB13 from membranes (PubMed:9712853). Interacts with prenylated INPP5E (PubMed:24166846). Interacts with RPGR (PubMed:9990021, PubMed:24166846, PubMed:23559067). Interacts with ARL2 (PubMed:24166846, PubMed:22002721). Interacts with ARL3; the interaction occurs specifically with the GTP-bound form of ARL3 (PubMed:24166846). Interaction with ARL2 and ARL3 promotes release of farnesylated cargo proteins (PubMed:22002721).
Similarity
Belongs to the PDE6D/unc-119 family.
Keywords
Cell projection
cGMP
Complete proteome
Cytoplasm
Cytoplasmic vesicle
Cytoskeleton
Membrane
Reference proteome
Sensory transduction
Vision
Direct protein sequencing
3D-structure
Ciliopathy
Joubert syndrome
Feature
chain Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta
Uniprot
A0A2A4JCP5
A0A2W1BL10
A0A2H1VGT6
S4P836
H9IUE9
A0A194RSP0
+ More
A0A222AJF3 A0A0N0BJV9 A0A3L8DRK9 A0A194PD65 F4X519 A0A154P3N7 E2ALH0 E2C3E8 A0A088AMF8 R4WDD2 A0A0F7VHB0 A0A1B6E4X0 E9ICR9 A0A087U942 A0A0A9Z326 A0A0V0GDB8 T1HZU2 A0A0K8R3M0 E0VSJ1 A0A224Z7U2 A0A131YJV4 A0A131XN42 A0A2B4SA18 A0A131Y355 A0A224XYW4 H3AW39 E2J7D6 A0A293MXS2 A0A1Z5L0H9 A7SFK7 A0A1U7RQZ7 K4FYN5 A0A0P7UZU8 U3F978 G9KFR8 A0A1W5ALY7 M3Y0G7 Q9XT54 H0ZHQ2 W5LZK1 A0A3Q2XNJ1 F1SMV2 Q6DH83 A0A3B3D8M1 F1QZ52 I3M7F1 K9IYB0 A0A3Q7QTA1 A0A2Y9H2Z7 A0A2U3XS88 A0A2I2V1R8 A0A2Y9IWV0 A0A2U3VM51 A0A3Q7UGI7 Q95142 G3RCF1 A0A2R9ACJ4 A0A2K5C118 A0A2K6NXU1 A0A2K5R3A5 A0A2K6TJP9 H2P8W8 H2QJL5 Q6IB24 O43924 A0A341B407 A0A2U3UYK7 A0A340XTZ8 A0A2Y9FJH4 A0A2Y9MN01 A0A384ABW9 A0A212EY25 A0A0P6J443 A0A023GBU3 A0A2Y9E9N8 A0A1B6KXW7 A0A1U7QZ77 A0A1B6G5Q2 A0A218UWD8 A0A3M0JMF3 A0A1V4KP47 U3JYV3 A0A2K6BU11 G7PKA4 A0A2K5YLR0 F7BMR1 W5U688 C3ZYY6 A0A3Q2CXT1 A0A1U7UZ61 H9GII4 V4BQV6 A0A3B3HQE9 A0A3P9ITK7
A0A222AJF3 A0A0N0BJV9 A0A3L8DRK9 A0A194PD65 F4X519 A0A154P3N7 E2ALH0 E2C3E8 A0A088AMF8 R4WDD2 A0A0F7VHB0 A0A1B6E4X0 E9ICR9 A0A087U942 A0A0A9Z326 A0A0V0GDB8 T1HZU2 A0A0K8R3M0 E0VSJ1 A0A224Z7U2 A0A131YJV4 A0A131XN42 A0A2B4SA18 A0A131Y355 A0A224XYW4 H3AW39 E2J7D6 A0A293MXS2 A0A1Z5L0H9 A7SFK7 A0A1U7RQZ7 K4FYN5 A0A0P7UZU8 U3F978 G9KFR8 A0A1W5ALY7 M3Y0G7 Q9XT54 H0ZHQ2 W5LZK1 A0A3Q2XNJ1 F1SMV2 Q6DH83 A0A3B3D8M1 F1QZ52 I3M7F1 K9IYB0 A0A3Q7QTA1 A0A2Y9H2Z7 A0A2U3XS88 A0A2I2V1R8 A0A2Y9IWV0 A0A2U3VM51 A0A3Q7UGI7 Q95142 G3RCF1 A0A2R9ACJ4 A0A2K5C118 A0A2K6NXU1 A0A2K5R3A5 A0A2K6TJP9 H2P8W8 H2QJL5 Q6IB24 O43924 A0A341B407 A0A2U3UYK7 A0A340XTZ8 A0A2Y9FJH4 A0A2Y9MN01 A0A384ABW9 A0A212EY25 A0A0P6J443 A0A023GBU3 A0A2Y9E9N8 A0A1B6KXW7 A0A1U7QZ77 A0A1B6G5Q2 A0A218UWD8 A0A3M0JMF3 A0A1V4KP47 U3JYV3 A0A2K6BU11 G7PKA4 A0A2K5YLR0 F7BMR1 W5U688 C3ZYY6 A0A3Q2CXT1 A0A1U7UZ61 H9GII4 V4BQV6 A0A3B3HQE9 A0A3P9ITK7
Pubmed
28756777
23622113
19121390
26354079
30249741
21719571
+ More
20798317 23691247 21282665 25401762 26823975 20566863 28797301 26830274 28049606 9215903 28528879 17615350 23056606 25243066 23236062 10452952 20360741 30723633 29451363 23594743 17975172 8798640 14561760 22398555 22722832 25362486 16136131 11181995 9570951 9533031 9781033 9712853 15489334 10518933 9990021 21269460 24166846 11980706 22002721 23559067 23698361 22118469 22002653 17431167 25319552 23127152 18563158 21881562 23254933 17554307
20798317 23691247 21282665 25401762 26823975 20566863 28797301 26830274 28049606 9215903 28528879 17615350 23056606 25243066 23236062 10452952 20360741 30723633 29451363 23594743 17975172 8798640 14561760 22398555 22722832 25362486 16136131 11181995 9570951 9533031 9781033 9712853 15489334 10518933 9990021 21269460 24166846 11980706 22002721 23559067 23698361 22118469 22002653 17431167 25319552 23127152 18563158 21881562 23254933 17554307
EMBL
NWSH01001942
PCG69619.1
KZ149983
PZC75762.1
ODYU01002500
SOQ40063.1
+ More
GAIX01006116 JAA86444.1 BABH01001171 KQ459700 KPJ20409.1 KY609772 ASO76487.1 KQ435711 KOX79598.1 QOIP01000005 RLU22936.1 KQ459606 KPI91177.1 GL888680 EGI58476.1 KQ434796 KZC05848.1 GL440607 EFN65728.1 GL452320 EFN77486.1 AK417573 BAN20788.1 LN830654 CFW94163.1 GEDC01021564 GEDC01004327 JAS15734.1 JAS32971.1 GL762354 EFZ21653.1 KK118801 KFM73881.1 GBHO01028419 GBHO01003972 GDHC01003710 JAG15185.1 JAG39632.1 JAQ14919.1 GECL01000028 JAP06096.1 ACPB03011807 GADI01008197 JAA65611.1 DS235751 EEB16347.1 GFPF01011306 MAA22452.1 GEDV01008988 JAP79569.1 GEFH01001033 JAP67548.1 LSMT01000119 PFX26731.1 GEFM01002881 JAP72915.1 GFTR01002644 JAW13782.1 AFYH01083657 AFYH01083658 AFYH01083659 HP429354 ADN29854.1 GFWV01020000 MAA44728.1 GFJQ02006083 JAW00887.1 DS469645 EDO37484.1 JX053274 AFK11502.1 JARO02005050 KPP67434.1 GAMT01008318 GAMS01002761 GAMR01008873 GAMQ01006195 GAMP01000690 JAB03543.1 JAB20375.1 JAB25059.1 JAB35656.1 JAB52065.1 JP015145 AES03743.1 AEYP01081359 AEYP01081360 AEYP01081361 AEYP01081362 AEYP01081363 AEYP01081364 AF113996 AF109151 AY029186 AJ427395 AJ427396 ABQF01023427 ABQF01023428 ABQF01023429 AHAT01022428 AHAT01022429 AEMK02000101 DQIR01223868 HDB79345.1 BC076096 AAH76096.1 CU459069 AGTP01044234 AGTP01044235 AGTP01044236 AGTP01044237 GABZ01008361 JAA45164.1 AANG04002752 U65073 CABD030019118 CABD030019119 CABD030019120 CABD030019121 CABD030019122 AJFE02075470 AJFE02075471 ABGA01277448 ABGA01277449 ABGA01277450 ABGA01277451 ABGA01277452 NDHI03003487 PNJ35200.1 AACZ04056637 AACZ04056638 GABC01010029 GABF01004999 GABD01005728 GABE01003100 NBAG03000230 JAA01309.1 JAA17146.1 JAA27372.1 JAA41639.1 PNI70378.1 AC073476 CR456980 CH471063 AAY24151.1 CAG33261.1 EAW70978.1 AF045999 AF022912 AF042835 AF042833 AF042834 AJ001626 BT007278 BC007831 AGBW02011630 OWR46396.1 GEBF01005494 JAN98138.1 GBBM01004056 JAC31362.1 GEBQ01023688 JAT16289.1 GECZ01012005 JAS57764.1 MUZQ01000110 OWK58019.1 QRBI01000134 RMC02049.1 LSYS01002834 OPJ85567.1 AGTO01020409 CM001287 EHH55245.1 JSUE03009833 JSUE03009834 JSUE03009835 JU334901 JU472104 JV045081 CM001264 AFE78654.1 AFH28908.1 AFI35152.1 EHH21758.1 JT406210 AHH37501.1 GG666738 EEN42257.1 AAWZ02024090 KB202283 ESO91294.1
GAIX01006116 JAA86444.1 BABH01001171 KQ459700 KPJ20409.1 KY609772 ASO76487.1 KQ435711 KOX79598.1 QOIP01000005 RLU22936.1 KQ459606 KPI91177.1 GL888680 EGI58476.1 KQ434796 KZC05848.1 GL440607 EFN65728.1 GL452320 EFN77486.1 AK417573 BAN20788.1 LN830654 CFW94163.1 GEDC01021564 GEDC01004327 JAS15734.1 JAS32971.1 GL762354 EFZ21653.1 KK118801 KFM73881.1 GBHO01028419 GBHO01003972 GDHC01003710 JAG15185.1 JAG39632.1 JAQ14919.1 GECL01000028 JAP06096.1 ACPB03011807 GADI01008197 JAA65611.1 DS235751 EEB16347.1 GFPF01011306 MAA22452.1 GEDV01008988 JAP79569.1 GEFH01001033 JAP67548.1 LSMT01000119 PFX26731.1 GEFM01002881 JAP72915.1 GFTR01002644 JAW13782.1 AFYH01083657 AFYH01083658 AFYH01083659 HP429354 ADN29854.1 GFWV01020000 MAA44728.1 GFJQ02006083 JAW00887.1 DS469645 EDO37484.1 JX053274 AFK11502.1 JARO02005050 KPP67434.1 GAMT01008318 GAMS01002761 GAMR01008873 GAMQ01006195 GAMP01000690 JAB03543.1 JAB20375.1 JAB25059.1 JAB35656.1 JAB52065.1 JP015145 AES03743.1 AEYP01081359 AEYP01081360 AEYP01081361 AEYP01081362 AEYP01081363 AEYP01081364 AF113996 AF109151 AY029186 AJ427395 AJ427396 ABQF01023427 ABQF01023428 ABQF01023429 AHAT01022428 AHAT01022429 AEMK02000101 DQIR01223868 HDB79345.1 BC076096 AAH76096.1 CU459069 AGTP01044234 AGTP01044235 AGTP01044236 AGTP01044237 GABZ01008361 JAA45164.1 AANG04002752 U65073 CABD030019118 CABD030019119 CABD030019120 CABD030019121 CABD030019122 AJFE02075470 AJFE02075471 ABGA01277448 ABGA01277449 ABGA01277450 ABGA01277451 ABGA01277452 NDHI03003487 PNJ35200.1 AACZ04056637 AACZ04056638 GABC01010029 GABF01004999 GABD01005728 GABE01003100 NBAG03000230 JAA01309.1 JAA17146.1 JAA27372.1 JAA41639.1 PNI70378.1 AC073476 CR456980 CH471063 AAY24151.1 CAG33261.1 EAW70978.1 AF045999 AF022912 AF042835 AF042833 AF042834 AJ001626 BT007278 BC007831 AGBW02011630 OWR46396.1 GEBF01005494 JAN98138.1 GBBM01004056 JAC31362.1 GEBQ01023688 JAT16289.1 GECZ01012005 JAS57764.1 MUZQ01000110 OWK58019.1 QRBI01000134 RMC02049.1 LSYS01002834 OPJ85567.1 AGTO01020409 CM001287 EHH55245.1 JSUE03009833 JSUE03009834 JSUE03009835 JU334901 JU472104 JV045081 CM001264 AFE78654.1 AFH28908.1 AFI35152.1 EHH21758.1 JT406210 AHH37501.1 GG666738 EEN42257.1 AAWZ02024090 KB202283 ESO91294.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000053105
UP000279307
UP000053268
+ More
UP000007755 UP000076502 UP000000311 UP000008237 UP000005203 UP000054359 UP000015103 UP000009046 UP000225706 UP000008672 UP000001593 UP000189705 UP000034805 UP000192224 UP000000715 UP000002254 UP000007754 UP000018468 UP000264820 UP000008227 UP000261560 UP000000437 UP000005215 UP000286641 UP000248481 UP000245341 UP000011712 UP000248482 UP000245340 UP000286642 UP000009136 UP000001519 UP000240080 UP000233020 UP000233200 UP000233040 UP000233220 UP000001595 UP000002277 UP000005640 UP000252040 UP000245320 UP000265300 UP000248484 UP000248483 UP000261681 UP000007151 UP000248480 UP000189706 UP000197619 UP000269221 UP000190648 UP000016665 UP000233120 UP000009130 UP000233140 UP000006718 UP000221080 UP000001554 UP000265020 UP000189704 UP000001646 UP000030746 UP000001038 UP000265180 UP000265200
UP000007755 UP000076502 UP000000311 UP000008237 UP000005203 UP000054359 UP000015103 UP000009046 UP000225706 UP000008672 UP000001593 UP000189705 UP000034805 UP000192224 UP000000715 UP000002254 UP000007754 UP000018468 UP000264820 UP000008227 UP000261560 UP000000437 UP000005215 UP000286641 UP000248481 UP000245341 UP000011712 UP000248482 UP000245340 UP000286642 UP000009136 UP000001519 UP000240080 UP000233020 UP000233200 UP000233040 UP000233220 UP000001595 UP000002277 UP000005640 UP000252040 UP000245320 UP000265300 UP000248484 UP000248483 UP000261681 UP000007151 UP000248480 UP000189706 UP000197619 UP000269221 UP000190648 UP000016665 UP000233120 UP000009130 UP000233140 UP000006718 UP000221080 UP000001554 UP000265020 UP000189704 UP000001646 UP000030746 UP000001038 UP000265180 UP000265200
PRIDE
Pfam
PF05351 GMP_PDE_delta
Interpro
SUPFAM
SSF81296
SSF81296
Gene 3D
ProteinModelPortal
A0A2A4JCP5
A0A2W1BL10
A0A2H1VGT6
S4P836
H9IUE9
A0A194RSP0
+ More
A0A222AJF3 A0A0N0BJV9 A0A3L8DRK9 A0A194PD65 F4X519 A0A154P3N7 E2ALH0 E2C3E8 A0A088AMF8 R4WDD2 A0A0F7VHB0 A0A1B6E4X0 E9ICR9 A0A087U942 A0A0A9Z326 A0A0V0GDB8 T1HZU2 A0A0K8R3M0 E0VSJ1 A0A224Z7U2 A0A131YJV4 A0A131XN42 A0A2B4SA18 A0A131Y355 A0A224XYW4 H3AW39 E2J7D6 A0A293MXS2 A0A1Z5L0H9 A7SFK7 A0A1U7RQZ7 K4FYN5 A0A0P7UZU8 U3F978 G9KFR8 A0A1W5ALY7 M3Y0G7 Q9XT54 H0ZHQ2 W5LZK1 A0A3Q2XNJ1 F1SMV2 Q6DH83 A0A3B3D8M1 F1QZ52 I3M7F1 K9IYB0 A0A3Q7QTA1 A0A2Y9H2Z7 A0A2U3XS88 A0A2I2V1R8 A0A2Y9IWV0 A0A2U3VM51 A0A3Q7UGI7 Q95142 G3RCF1 A0A2R9ACJ4 A0A2K5C118 A0A2K6NXU1 A0A2K5R3A5 A0A2K6TJP9 H2P8W8 H2QJL5 Q6IB24 O43924 A0A341B407 A0A2U3UYK7 A0A340XTZ8 A0A2Y9FJH4 A0A2Y9MN01 A0A384ABW9 A0A212EY25 A0A0P6J443 A0A023GBU3 A0A2Y9E9N8 A0A1B6KXW7 A0A1U7QZ77 A0A1B6G5Q2 A0A218UWD8 A0A3M0JMF3 A0A1V4KP47 U3JYV3 A0A2K6BU11 G7PKA4 A0A2K5YLR0 F7BMR1 W5U688 C3ZYY6 A0A3Q2CXT1 A0A1U7UZ61 H9GII4 V4BQV6 A0A3B3HQE9 A0A3P9ITK7
A0A222AJF3 A0A0N0BJV9 A0A3L8DRK9 A0A194PD65 F4X519 A0A154P3N7 E2ALH0 E2C3E8 A0A088AMF8 R4WDD2 A0A0F7VHB0 A0A1B6E4X0 E9ICR9 A0A087U942 A0A0A9Z326 A0A0V0GDB8 T1HZU2 A0A0K8R3M0 E0VSJ1 A0A224Z7U2 A0A131YJV4 A0A131XN42 A0A2B4SA18 A0A131Y355 A0A224XYW4 H3AW39 E2J7D6 A0A293MXS2 A0A1Z5L0H9 A7SFK7 A0A1U7RQZ7 K4FYN5 A0A0P7UZU8 U3F978 G9KFR8 A0A1W5ALY7 M3Y0G7 Q9XT54 H0ZHQ2 W5LZK1 A0A3Q2XNJ1 F1SMV2 Q6DH83 A0A3B3D8M1 F1QZ52 I3M7F1 K9IYB0 A0A3Q7QTA1 A0A2Y9H2Z7 A0A2U3XS88 A0A2I2V1R8 A0A2Y9IWV0 A0A2U3VM51 A0A3Q7UGI7 Q95142 G3RCF1 A0A2R9ACJ4 A0A2K5C118 A0A2K6NXU1 A0A2K5R3A5 A0A2K6TJP9 H2P8W8 H2QJL5 Q6IB24 O43924 A0A341B407 A0A2U3UYK7 A0A340XTZ8 A0A2Y9FJH4 A0A2Y9MN01 A0A384ABW9 A0A212EY25 A0A0P6J443 A0A023GBU3 A0A2Y9E9N8 A0A1B6KXW7 A0A1U7QZ77 A0A1B6G5Q2 A0A218UWD8 A0A3M0JMF3 A0A1V4KP47 U3JYV3 A0A2K6BU11 G7PKA4 A0A2K5YLR0 F7BMR1 W5U688 C3ZYY6 A0A3Q2CXT1 A0A1U7UZ61 H9GII4 V4BQV6 A0A3B3HQE9 A0A3P9ITK7
PDB
5T4X
E-value=4.10108e-65,
Score=624
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytosol
Cytoplasmic vesicle membrane
Cytoskeleton
Cilium basal body
Cytoplasm
Cytosol
Cytoplasmic vesicle membrane
Cytoskeleton
Cilium basal body
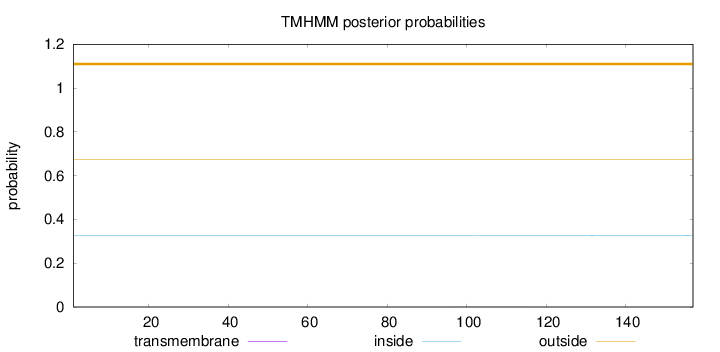
Length:
157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00544
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.32638
outside
1 - 157
Population Genetic Test Statistics
Pi
326.721839
Theta
218.182719
Tajima's D
1.583725
CLR
0.321505
CSRT
0.8029598520074
Interpretation
Uncertain
