Gene
KWMTBOMO07718
Pre Gene Modal
BGIBMGA000878
Annotation
PREDICTED:_uncharacterized_protein_LOC101742063_isoform_X1_[Bombyx_mori]
Full name
Pyruvate dehydrogenase E1 component subunit alpha
Location in the cell
Nuclear Reliability : 2.851
Sequence
CDS
ATGGAGCAAGTCATTCTAGTCTCTATAGACATTGATAACAATTTAATAACGGATAAATGTGTCTTGGACCAGGCCATGGACGATATTGAGTTCCTGGCTGACTACCCACAGTACACCGGCAACCGTCACATAGTTTGGCTATTCCTGATGGAGTTAGCGAGACGTCGCTGGGAGGCCAGACCAAGGGCAGAGCAAGAACGCGCTACACAACTATTGGAAGGCGCTGGACAACCATTTAAGGAAATGGAGAACGTTGTCTGGGAACAAAGAGTACACTTCGCAGCTGCCATCGCTAGGATTCGTATAAAAAATAAGGCTCTCTCGTTGTCGGACCTATTGCCGCCACATTTGCGTGAGGAACGCATATCAGCGTGTGTCAATAAGGACATAATCACGGGTTGGGTTAACACTTTTAAAGCCAAAAAAATTGCATTGCTGGTTAAAAAATTACACGAACTAGGATACTCTTACAGCAGCTCACGACAGCTTTGCGCCGGCGAATACAGATTTGATAGAGTTTGTCCGAGATTCATAACGCTACGACCTCCGGCCAGTATCAGCATTGGGAAATTGGATTTAGTACAAGAAGGAATTATTGTTCTTCAGGAACGCGAGTTTTGCGAGGGAGCTTCTACGCTTTGTCGAGCCCTTCGCGCGAACTCTCTTCAAGGAGTGGTGGCCCAGACCCACGCGTCGTCCCCCCGTTGCTCAGCGTACCTCGCGGCTCAGCTGAAGGAACTGGCCGCAGTGCTGAAGGCCCAGACTCCTCCTGCACCAGCTATACCGGAACTCGGTCGACTGGTTGTGTTCGGAGCAGGAGATAAAGCCGAAAGTTATGTTTTAGCTCTTCGGGAGCTCGGCATAGAGGCCTCAGAGGCGCCTCACTCGGGAGCGCCAGTATGCGTCCTTAGCGAACCTGTACATTGCGACACTCCGACTGTGACCAACGCGTTAGACGGAGTGGTAGCAGCCTTGGCAACTCCACCCAACTCTTACTCCGCAGTCACGGATCCCATTGATCTTGTGTGCGGTCGTGGAGGAGATTTGGCAATGCTAGAGGTTTTAACCGAATCGGAGATTGACAGTAAAGGTAAAGCTAGAGTGCAATCAATTCTCGAGGAACAAAAGAAAACATTGAAGACCTTAATGTCTAAGCCTCAAATCCAGTTGATATTGTACGAAACACATTCTGCATTAGAAGCCGAAAATCAAGCGCAAGTGACAAGAGCCGTCGCAGAAGCGAACAGATTGGCTCGGGAACGACACGCGTTGCTGAAAAGGAAACATCGCCAATCCAGTCCATTAAAGCACGGAATAGAAAACATTGAGTCCTTAACAACATTAGACAACACAATCTCTGAGGGAGACAGAGATGACCAAACGGATGATGAATTTGATTCAATTTCACCAAAGCGACGCACGTCTGGACCGTCGCCGCCAAGACGGAGATCTCAGACCGACGATACAGTGTTAAGAAAACAAGCGGATTCATCTCGTCCCGGATCAAGGCCTGGGACTAACAAAGCCCGACCTATACCTGCAGTACCCGATGACGCCGTCCCCGGAGATCCCGACAAGGATAATCCGGATATTTTTGTACCGAATTGCGATTTGTTCGAAATCGCGACGTTGCCTAATTTAGGGAATGGTTTAGATATTAACTACATACTGGATCGAGACGGTTGTTATCTCGGCCTCATCCAAAGGAAGGAGATAACCCGTTTGGACGCAAAGTATATGATCCGTATGGCGGAGGAGCGTGGGCTTTTCGGCGCGAGCGCGGCCGCCGCGTCCCCCCCGCGGCGGAGGCGCGCCGACACCGCGCCCTCGCGACACAAGCGGGCCCGCAGGAGAACCAGCTTTGAGGTAGAACGTATAGCCGCTCCGACGTACGCGTCGCTGTGCCGATCGTCGCGTCGCGGCAGCGCGCCTGCCTGTCTACAAGACGCGCCCCCCGAGCAACAGTTGCTCGAGTGTCAGCGGCACGCGCGGCGACAGGCGGCCGCGGCGGCAATTACTGCCGCGGCCTGTAACTGTGACGCCAGGCACAGAACGCCATACCGTCGGGCATCAGCTGATGTGACGGTCACAGCGCGACCCGCCTCGCCCCCGTGCCGACGACCCTTCCCACTGGTAGTACACGATCTTAGACTCAAGTCTATACCTATGACTACCCACAAAATATTCGCTTGA
Protein
MEQVILVSIDIDNNLITDKCVLDQAMDDIEFLADYPQYTGNRHIVWLFLMELARRRWEARPRAEQERATQLLEGAGQPFKEMENVVWEQRVHFAAAIARIRIKNKALSLSDLLPPHLREERISACVNKDIITGWVNTFKAKKIALLVKKLHELGYSYSSSRQLCAGEYRFDRVCPRFITLRPPASISIGKLDLVQEGIIVLQEREFCEGASTLCRALRANSLQGVVAQTHASSPRCSAYLAAQLKELAAVLKAQTPPAPAIPELGRLVVFGAGDKAESYVLALRELGIEASEAPHSGAPVCVLSEPVHCDTPTVTNALDGVVAALATPPNSYSAVTDPIDLVCGRGGDLAMLEVLTESEIDSKGKARVQSILEEQKKTLKTLMSKPQIQLILYETHSALEAENQAQVTRAVAEANRLARERHALLKRKHRQSSPLKHGIENIESLTTLDNTISEGDRDDQTDDEFDSISPKRRTSGPSPPRRRSQTDDTVLRKQADSSRPGSRPGTNKARPIPAVPDDAVPGDPDKDNPDIFVPNCDLFEIATLPNLGNGLDINYILDRDGCYLGLIQRKEITRLDAKYMIRMAEERGLFGASAAAASPPRRRRADTAPSRHKRARRRTSFEVERIAAPTYASLCRSSRRGSAPACLQDAPPEQQLLECQRHARRQAAAAAITAAACNCDARHRTPYRRASADVTVTARPASPPCRRPFPLVVHDLRLKSIPMTTHKIFA
Summary
Description
The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2).
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
[dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-lipoyl-L-lysine + H(+) + pyruvate = [dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-(S(8)-acetyldihydrolipoyl)-L-lysine + CO2
[dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-lipoyl-L-lysine + H(+) + pyruvate = [dihydrolipoyllysine-residue acetyltransferase]-(R)-N(6)-(S(8)-acetyldihydrolipoyl)-L-lysine + CO2
Cofactor
thiamine diphosphate
Uniprot
A0A2W1BQ72
A0A2H1VGU1
A0A194PEP8
A0A194RSU1
A0A2A4JXD9
A0A212EY68
+ More
A0A0L7L6H2 A0A1Y1L1J0 A0A1Y1KWR6 A0A1Y1L414 A0A2J7PZE4 A0A2J7PZE5 A0A2J7PZE7 A0A2J7PZE6 A0A1W4XEB5 A0A336MCW3 A0A336M9L1 A0A182M5F0 A0A182WZQ6 A0A2C9GQX1 A0A182U1B0 A0A182VCJ2 Q7Q0B2 A0A084WIE2 A0A182JNV3 A0A182WG36 A0A1Y9HEP9 W5JW12 Q29EI6 A0A182FB97 A0A0R3P940 N6UDB2 A0A151JP29 A0A151X9U6 A0A195CV24
A0A0L7L6H2 A0A1Y1L1J0 A0A1Y1KWR6 A0A1Y1L414 A0A2J7PZE4 A0A2J7PZE5 A0A2J7PZE7 A0A2J7PZE6 A0A1W4XEB5 A0A336MCW3 A0A336M9L1 A0A182M5F0 A0A182WZQ6 A0A2C9GQX1 A0A182U1B0 A0A182VCJ2 Q7Q0B2 A0A084WIE2 A0A182JNV3 A0A182WG36 A0A1Y9HEP9 W5JW12 Q29EI6 A0A182FB97 A0A0R3P940 N6UDB2 A0A151JP29 A0A151X9U6 A0A195CV24
EC Number
1.2.4.1
Pubmed
EMBL
KZ149983
PZC75764.1
ODYU01002500
SOQ40065.1
KQ459606
KPI91169.1
+ More
KQ459700 KPJ20415.1 NWSH01000400 PCG76681.1 AGBW02011630 OWR46394.1 JTDY01002656 KOB71000.1 GEZM01071410 JAV65815.1 GEZM01071411 JAV65814.1 GEZM01071409 GEZM01071405 JAV65817.1 NEVH01020335 PNF21708.1 PNF21706.1 PNF21705.1 PNF21707.1 UFQT01000782 SSX27191.1 UFQS01000758 UFQT01000758 SSX06688.1 SSX27034.1 AXCM01004871 APCN01003303 AAAB01008986 EAA00406.4 ATLV01023929 KE525347 KFB49986.1 ADMH02000120 ETN67688.1 CH379070 EAL30074.3 KRT08630.1 APGK01039344 APGK01039345 APGK01039346 APGK01039347 APGK01039348 KB740970 ENN76642.1 KQ978820 KYN28138.1 KQ982365 KYQ57133.1 KQ977279 KYN04377.1
KQ459700 KPJ20415.1 NWSH01000400 PCG76681.1 AGBW02011630 OWR46394.1 JTDY01002656 KOB71000.1 GEZM01071410 JAV65815.1 GEZM01071411 JAV65814.1 GEZM01071409 GEZM01071405 JAV65817.1 NEVH01020335 PNF21708.1 PNF21706.1 PNF21705.1 PNF21707.1 UFQT01000782 SSX27191.1 UFQS01000758 UFQT01000758 SSX06688.1 SSX27034.1 AXCM01004871 APCN01003303 AAAB01008986 EAA00406.4 ATLV01023929 KE525347 KFB49986.1 ADMH02000120 ETN67688.1 CH379070 EAL30074.3 KRT08630.1 APGK01039344 APGK01039345 APGK01039346 APGK01039347 APGK01039348 KB740970 ENN76642.1 KQ978820 KYN28138.1 KQ982365 KYQ57133.1 KQ977279 KYN04377.1
Proteomes
Interpro
CDD
ProteinModelPortal
A0A2W1BQ72
A0A2H1VGU1
A0A194PEP8
A0A194RSU1
A0A2A4JXD9
A0A212EY68
+ More
A0A0L7L6H2 A0A1Y1L1J0 A0A1Y1KWR6 A0A1Y1L414 A0A2J7PZE4 A0A2J7PZE5 A0A2J7PZE7 A0A2J7PZE6 A0A1W4XEB5 A0A336MCW3 A0A336M9L1 A0A182M5F0 A0A182WZQ6 A0A2C9GQX1 A0A182U1B0 A0A182VCJ2 Q7Q0B2 A0A084WIE2 A0A182JNV3 A0A182WG36 A0A1Y9HEP9 W5JW12 Q29EI6 A0A182FB97 A0A0R3P940 N6UDB2 A0A151JP29 A0A151X9U6 A0A195CV24
A0A0L7L6H2 A0A1Y1L1J0 A0A1Y1KWR6 A0A1Y1L414 A0A2J7PZE4 A0A2J7PZE5 A0A2J7PZE7 A0A2J7PZE6 A0A1W4XEB5 A0A336MCW3 A0A336M9L1 A0A182M5F0 A0A182WZQ6 A0A2C9GQX1 A0A182U1B0 A0A182VCJ2 Q7Q0B2 A0A084WIE2 A0A182JNV3 A0A182WG36 A0A1Y9HEP9 W5JW12 Q29EI6 A0A182FB97 A0A0R3P940 N6UDB2 A0A151JP29 A0A151X9U6 A0A195CV24
Ontologies
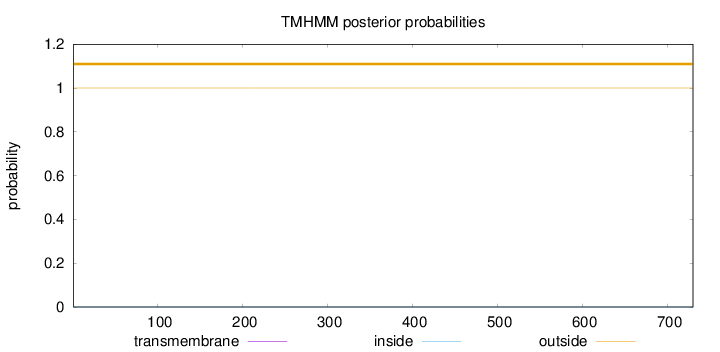
Topology
Length:
730
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00049
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00008
outside
1 - 730
Population Genetic Test Statistics
Pi
212.836716
Theta
155.233257
Tajima's D
1.50258
CLR
0.30449
CSRT
0.783360831958402
Interpretation
Uncertain
