Gene
KWMTBOMO07716
Pre Gene Modal
BGIBMGA001268
Annotation
PREDICTED:_phosphatidylinositol_4-phosphate_3-kinase_C2_domain-containing_subunit_alpha_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.585
Sequence
CDS
ATGTCTGACTACGATCTTCAGTTTCAAGCGGATTTAGAAAAGGCGCAGGCTTTGAGTTTAGAAAGTCTCGCGCTAGAACAATTTCGAAGGCAGAAGCTTCTTTCTAGCACAGATAATGGACGGGAGTTGGGCACTGTCAATTATCGTCGCAGAGAGGACGAGGGGCCCCCGCCAGCGCTGCCGCCACCTCCGCGGAAACCACCTCGTGGACGATCCAATTCCGAAACTCCAAGGTCATCTGAGGCGAGTCCCACCAGGGACGAGAGCAGCGACCTGATCTCGTTTGCAAGTCCAACCAGCAAGGCAGCACCGCAAGATCCACATGCGTCACTCAAAGAATATATAGACCAGATCAACAAACTAGGTCAAAAAACTGAAGACTTGACAGAACAATCGCCTGTAATCATGAATAATCATCCGTACAACCAAAGGCCCGGTTATCCTTACGCGCCGCAATACAACTACAACTATCCCCCTTACAACGTGATGCCTCCATATCCGTCTTCCGCGCCCTACGGCATGCCATCACCCATGCCGAATTATTCGTATTACACGCCGCCCATAGCTGCACCACCGGAGACGCCGCCCGCATACGTGCCAGGGAACCCGGCGATGTATTCCGTTCCATACAGCCAAACGCGGATGTCAGCTCCCGTCACTCCCAACTATGGATATCCACCGGCGTACAATTACAACACGACGAACATGATATCTCGTCCGCCGATGCCTATTTCGAACTTGTACCCGCAGCTGAGTCAGGCCCACGCTTACCCGCCGAACTCGAACCAGCCGCAAAACAAAAGCAACGACACCACACAGACGATCGTTAAAACGGAACCGTCAAGGGACAGCATGGACAGTAATAGCTCGAGGAAATCGTCTGAAGTTAGCATAGGGTCCAACGAATCAAACGACACAGTAGGAAGTCTCCCTAAATCAATGAACCTGATCGACTTTGGTGGTCGAAACCCGAACGGCAGTCAAGAATATGCGGTGGGCAGCGAATGTTCGAGTAGCGTCTACGAAGAGTACGACCCGCTGGATTTCCTGTACGGGGAGACTGAGATGCGTGAGACGCCCGTATACGGTACCGTCGGCAAGCGCGAGCCTGCCGTCATGCCGTCGCCGCCGCCAAAGGCAATGCCCTCCGTGCCCGCCGTCGCGCTCAGATCCACCACGAACAAACTTTACGATAGCATAGTTAAGATCAGAACAAGAAATTACGACAGCGAACACAAAGCGTTTCTTAAAATGGCTCTTGACGTTCGAAAAAAGTTCGTGTACACTGATGAGAGGACGAACCCAGGGCACGTCATAGCAGCCAGGTCCGGCGGCAAGTACCCGCCCAACACGAGCATCAAGATACTAGTTCATCACGCTCACTCCGATGAACCCATCACTTTCACGTCCGATGTGGAATCTAAAGTCGAACACGTGATATTAACAGTAGTCTGTTCACTGGATGAAGACATCAGGGAGGACATGTCTGGATATATGTTAAGGGTGCGGGGAGCCAAAGAGTATCTGACGCCCGGCAGCTCGCTGAGCGAATACGATTACATACGCGAGTGCGTGCGCTTGGAGAAGGACGTGACGCTGTGTCTGCACGAAGGGAGGGACAGCACGCTGGCCAGGACCGAGAGGGACGACGCGAAGGACTCGCACCTCACGCTCGACGACCTGTTCCCGGCGCAGGGAGCCACGCCGATGCTATCGTTCGATAATTTATCGATATTGCTCGAAACTTTAGAAGGCGAGCTGTCGCGTGCTATCGGTGTGACCAGCGCTGAGGGCAGCTGCTCCCCGAAGGCGGTGTTCCAAGCCGTCAAGGCTATCTGCGCTCTGGCGGGCGGCGTGGAGACTCTGCACGTCACTCACACGCTCAACGCGTTCGCGCAGGCCTGCACCGCGCAGGGAGCGAACAACGTGGTCCGCCCGGAGATAGAGCGCGAACAGGGCGCGTACTGCTCGGTGTCGCTGGCGGCGGGCTCGCGGCGCGACTGCGTGTCGCTGCAGGGCGGGCGCGTGCGCGACGCCGTGCGGGCGCTGCTCGCGCTCTATACGCGCACGCGCCTGCTCGACTTCCGCCTCGCCGACGCGCACTACCAGGCGCCCGCCAACCCCGCCAAAGCAGTACCGGCGCACACAATATCAGAAACGACACTGTTTTGTATAGAGGCGGTGCACTGTTTGCCTCTGTGCTGGAGCCACGACGAGTACATGTTCCATGCGCAAATCTACCACGGAACAAGACCTTTGAAAACATTACGGTTCACACAAGCTTCGAAGATTGAATGCGGCGGGTTTTATCCTCGAATCATATTTGATACTTGGCTGAACCTCGAGGATTTACCAGTAAACACGTTACCAAGAGAATCTCGTCTCGTTCTGACGCTGCACGGCAGGACGCAGCGCACCATCGACAGCCAGCATCCGAACGAGAACAGCCCGCCCAGCAGCCAGGAGGCCGAGGACAGCGGCGACGTGCTGTACGAGCAGGTCGAGCTGGGCTGGGCCGCCCTGCAACTCTTCGACTTTGATGGGATGCTATCGTCCGGTAATTACGTTTTACCATTGTGGCCGACTAGCTGCGACCGGCGGACCGGCCCGGCTCCTCATCCCATGCACACACCTACAACACCATATCCCGTTGTCAATATTGAAATTCCTTTATACGCACACACCGGGGTCAAGTGGACCAGCGGAGAAGAAGACAAACAGAAGAATATATCTCTGAAGGATCTTCCATCGTTCGACAGTTTGGATGGACACACACAGTCTCAACTGCTACACCTCATCGAGCAGGGCGCTTATCAGAGGATGCCTACAGAATGTAGAGAGGTCATGTGGGAGAAACGGCAGTACCTGGTACAGATCCCAGGCGCTTTGCCTCTGGTGTTGCTAGCAGCTACAAATTGGTACGGAGACCATAGAGAACAATTGATAGGTCTACTCCGACTGTGGGAGAAACCTTCACCTTTAAACGCTATGCATTTGCTGTTGCCTTGTTTCCCGGATGTCGCCGTAAGAGAAACAGCAGCCAAGCTGTTGAACGAGATGTCTGACGACGACCTGTGCCGCGTGTTGCCGCAGCTCACGCAGGCGCTGCGACACGAGACGTACGAACCCAGTCCTTTAGCCGAGCTACTCCTCTCGCGAGCGCTAGCGGCTCCGGCTGTTGCTCACAGACTGTTTTGGCTTTTAGTGCATTCCCTGCCTAATATACCACAGGCGCCACATCAAGAGTTCCTCGGGATAAATTTAGAAGATAATGCTTCTGAAAACGCCGAAACTGATGCATCGATGAGCGCCTCGTGGCGCTACACGCTGATGCTGCGCGCCTTACTCGTTATATGCGGAGAAAATCTCAACAAGACGCTGCTTCGACAACAGTTACTCGTCAAGAGTCTGTGCGAGATGGCGGTGTCGGTGAAGTGCGCGAAGGAGTGCGCCCGCGGACGCGCGCTGTGTGCGGGCGGCACGGCGCTGGCGGGCGCGCTGCGTGCCCAGCCCACCGCACTGCCTCTCGCACTCCATAAATACGTACACGATCTCGACGTCAAATCATGTACCTACTTCTCATCAAACACACTTCCGCTGAAAATAAATTTTATTGGAACAGATAATGCAGTAATTCCAGCTATGTTCAAGGTTGGAGATGATTTACGACAAGATGCTCTAGTGTTGCAAGTGATAAAAATAATGGATACATTATGGTTGAAAGCTGGCTTAGATCTGAGGATGGTTACTTTCCAAGCTCTACCCACAAGTAATAGTAAAGGTATGATAGAAATAGTGACAGAAGCGGAGACGCTGCGGGCCATACAAACCGAGTGGGGTCTGACCGGCTCGTTCAAGGACAAGCCCATAGCCGAGTGGCTCGCCAAACACAATCCCTCCGAACTCGAATACCAACGCGCCAGAGACAATTTCACAGCTTCGTGCGCCGGGTACAGCGTGGCGACGTACCTGCTGGGGATCTGCGACCGGCACAACGACAACATCATGCTGAAGACCTCCGGCCACCTGTTCCACATCGACTTCGGGAAGTTCCTGGGAGACGCGCAAATGCAAACGAGCATACCGCCCACCTGA
Protein
MSDYDLQFQADLEKAQALSLESLALEQFRRQKLLSSTDNGRELGTVNYRRREDEGPPPALPPPPRKPPRGRSNSETPRSSEASPTRDESSDLISFASPTSKAAPQDPHASLKEYIDQINKLGQKTEDLTEQSPVIMNNHPYNQRPGYPYAPQYNYNYPPYNVMPPYPSSAPYGMPSPMPNYSYYTPPIAAPPETPPAYVPGNPAMYSVPYSQTRMSAPVTPNYGYPPAYNYNTTNMISRPPMPISNLYPQLSQAHAYPPNSNQPQNKSNDTTQTIVKTEPSRDSMDSNSSRKSSEVSIGSNESNDTVGSLPKSMNLIDFGGRNPNGSQEYAVGSECSSSVYEEYDPLDFLYGETEMRETPVYGTVGKREPAVMPSPPPKAMPSVPAVALRSTTNKLYDSIVKIRTRNYDSEHKAFLKMALDVRKKFVYTDERTNPGHVIAARSGGKYPPNTSIKILVHHAHSDEPITFTSDVESKVEHVILTVVCSLDEDIREDMSGYMLRVRGAKEYLTPGSSLSEYDYIRECVRLEKDVTLCLHEGRDSTLARTERDDAKDSHLTLDDLFPAQGATPMLSFDNLSILLETLEGELSRAIGVTSAEGSCSPKAVFQAVKAICALAGGVETLHVTHTLNAFAQACTAQGANNVVRPEIEREQGAYCSVSLAAGSRRDCVSLQGGRVRDAVRALLALYTRTRLLDFRLADAHYQAPANPAKAVPAHTISETTLFCIEAVHCLPLCWSHDEYMFHAQIYHGTRPLKTLRFTQASKIECGGFYPRIIFDTWLNLEDLPVNTLPRESRLVLTLHGRTQRTIDSQHPNENSPPSSQEAEDSGDVLYEQVELGWAALQLFDFDGMLSSGNYVLPLWPTSCDRRTGPAPHPMHTPTTPYPVVNIEIPLYAHTGVKWTSGEEDKQKNISLKDLPSFDSLDGHTQSQLLHLIEQGAYQRMPTECREVMWEKRQYLVQIPGALPLVLLAATNWYGDHREQLIGLLRLWEKPSPLNAMHLLLPCFPDVAVRETAAKLLNEMSDDDLCRVLPQLTQALRHETYEPSPLAELLLSRALAAPAVAHRLFWLLVHSLPNIPQAPHQEFLGINLEDNASENAETDASMSASWRYTLMLRALLVICGENLNKTLLRQQLLVKSLCEMAVSVKCAKECARGRALCAGGTALAGALRAQPTALPLALHKYVHDLDVKSCTYFSSNTLPLKINFIGTDNAVIPAMFKVGDDLRQDALVLQVIKIMDTLWLKAGLDLRMVTFQALPTSNSKGMIEIVTEAETLRAIQTEWGLTGSFKDKPIAEWLAKHNPSELEYQRARDNFTASCAGYSVATYLLGICDRHNDNIMLKTSGHLFHIDFGKFLGDAQMQTSIPPT
Summary
Similarity
Belongs to the PI3/PI4-kinase family.
Uniprot
H9IVI8
A0A2W1BRJ4
A0A194PJA3
A0A2H1VGX9
A0A212EY28
A0A2A4JZ40
+ More
A0A232FL80 K7IW81 A0A151IFN4 E2ABJ5 A0A1B6CT28 F4WQ66 A0A1B6D833 A0A195BML6 A0A151WKS4 A0A158NXE4 E9IVV7 A0A195ECF4 A0A0J7NVG0 A0A195FG44 A0A154PSM0 A0A3L8E4C0 E2BVU7 A0A2P8ZDS3 A0A088AC41 A0A2A3EGV0 T1HF92 A0A310SA10 A0A0N0U690 A0A0L7RKB3 A0A0C9PVJ8 A0A0C9RM03 A0A023F2W8 E0VBF9 A0A1Y1L4Z2 A0A1L8DL86 A0A1B0CCJ7 W4VRG7 Q0IGF6 A0A182GNH9 A0NBA9 A0A1Q3F7H7 A0A1Q3F7G5 A0A182QRR1 B0W7X6 A0A182FTM6 A0A2M4B8C5 A0A336MPB8 A0A2M3YZF9 A0A336LRK7 J9JP72 A0A2M4CHI7 A0A2M4CHG7 A0A2M4CHI5 A0A2M4B8D1 A0A182RUL2 A0A2S2NPJ6 A0A182KKM4 A0A2M4B8A2 W5JEA5 A0A182V420 A0A182MQA5 A0A1A9YEL9 A0A0K8VEC8 A0A1B0B1B1 A0A1B0A2K1 A0A2M4A4J6 A0A1B0FDB1 A0A2M3Z0P3 A0A2M4A4C5 A0A2M4A4P5 A0A1A9VMZ6 A0A034V3S2 A0A182WDV0 A0A182N318 W8BAJ7 A0A182YBJ8 A0A182PSA9 A0A2M4A771 A0A2S2QGW6 A0A0A1WPZ0 A0A182JVT2 A0A1A9X0C7 A0A1I8M8C7 A0A1I8NM56 A0A0L0CCG5 A0A139WA97 A0A084VU08 A0A139WAA5 A0A182IQY2 A0A0Q9WM38 B4LCS5 T1PAR1
A0A232FL80 K7IW81 A0A151IFN4 E2ABJ5 A0A1B6CT28 F4WQ66 A0A1B6D833 A0A195BML6 A0A151WKS4 A0A158NXE4 E9IVV7 A0A195ECF4 A0A0J7NVG0 A0A195FG44 A0A154PSM0 A0A3L8E4C0 E2BVU7 A0A2P8ZDS3 A0A088AC41 A0A2A3EGV0 T1HF92 A0A310SA10 A0A0N0U690 A0A0L7RKB3 A0A0C9PVJ8 A0A0C9RM03 A0A023F2W8 E0VBF9 A0A1Y1L4Z2 A0A1L8DL86 A0A1B0CCJ7 W4VRG7 Q0IGF6 A0A182GNH9 A0NBA9 A0A1Q3F7H7 A0A1Q3F7G5 A0A182QRR1 B0W7X6 A0A182FTM6 A0A2M4B8C5 A0A336MPB8 A0A2M3YZF9 A0A336LRK7 J9JP72 A0A2M4CHI7 A0A2M4CHG7 A0A2M4CHI5 A0A2M4B8D1 A0A182RUL2 A0A2S2NPJ6 A0A182KKM4 A0A2M4B8A2 W5JEA5 A0A182V420 A0A182MQA5 A0A1A9YEL9 A0A0K8VEC8 A0A1B0B1B1 A0A1B0A2K1 A0A2M4A4J6 A0A1B0FDB1 A0A2M3Z0P3 A0A2M4A4C5 A0A2M4A4P5 A0A1A9VMZ6 A0A034V3S2 A0A182WDV0 A0A182N318 W8BAJ7 A0A182YBJ8 A0A182PSA9 A0A2M4A771 A0A2S2QGW6 A0A0A1WPZ0 A0A182JVT2 A0A1A9X0C7 A0A1I8M8C7 A0A1I8NM56 A0A0L0CCG5 A0A139WA97 A0A084VU08 A0A139WAA5 A0A182IQY2 A0A0Q9WM38 B4LCS5 T1PAR1
Pubmed
EMBL
BABH01001177
BABH01001178
BABH01001179
BABH01001180
BABH01001181
KZ149983
+ More
PZC75767.1 KQ459606 KPI91165.1 ODYU01002500 SOQ40067.1 AGBW02011630 OWR46390.1 NWSH01000400 PCG76680.1 NNAY01000053 OXU31504.1 KQ977780 KYM99950.1 GL438292 EFN69190.1 GEDC01020678 GEDC01012952 JAS16620.1 JAS24346.1 GL888262 EGI63689.1 GEDC01029021 GEDC01015563 GEDC01005018 JAS08277.1 JAS21735.1 JAS32280.1 KQ976441 KYM86393.1 KQ983001 KYQ48479.1 ADTU01003040 ADTU01003041 GL766449 EFZ15298.1 KQ979074 KYN22898.1 LBMM01001404 KMQ96380.1 KQ981610 KYN39363.1 KQ435053 KZC14258.1 QOIP01000001 RLU27580.1 GL451029 EFN80176.1 PYGN01000086 PSN54649.1 KZ288258 PBC30512.1 ACPB03015469 ACPB03015470 ACPB03015471 KQ762659 OAD55550.1 KQ435735 KOX77152.1 KQ414574 KOC71253.1 GBYB01005478 JAG75245.1 GBYB01014407 JAG84174.1 GBBI01003159 JAC15553.1 DS235030 EEB10715.1 GEZM01070994 JAV66147.1 GFDF01006855 JAV07229.1 AJWK01006860 AJWK01006861 AJWK01006862 GANO01003076 JAB56795.1 CH477192 EAT48581.1 JXUM01000533 JXUM01000534 AAAB01008804 EAU77730.2 GFDL01011566 JAV23479.1 GFDL01011576 JAV23469.1 AXCN02001143 DS231856 EDS38438.1 GGFJ01000145 MBW49286.1 UFQT01001941 SSX32186.1 GGFM01000903 MBW21654.1 UFQS01000052 UFQT01000052 SSW98661.1 SSX19047.1 ABLF02039321 ABLF02039331 GGFL01000629 MBW64807.1 GGFL01000624 MBW64802.1 GGFL01000628 MBW64806.1 GGFJ01000146 MBW49287.1 GGMR01006057 MBY18676.1 GGFJ01000144 MBW49285.1 ADMH02001385 ETN62687.1 AXCM01006030 GDHF01015414 JAI36900.1 JXJN01007040 JXJN01007041 GGFK01002350 MBW35671.1 CCAG010006850 GGFM01001351 MBW22102.1 GGFK01002343 MBW35664.1 GGFK01002371 MBW35692.1 GAKP01022532 JAC36420.1 GAMC01019691 JAB86864.1 GGFK01003308 MBW36629.1 GGMS01007775 MBY76978.1 GBXI01013809 JAD00483.1 JRES01000610 KNC29902.1 KQ971410 KYB24843.1 ATLV01016620 KE525097 KFB41452.1 KYB24844.1 CH940647 KRF85222.1 EDW70967.1 KRF85218.1 KRF85219.1 KRF85220.1 KRF85221.1 KA645003 AFP59632.1
PZC75767.1 KQ459606 KPI91165.1 ODYU01002500 SOQ40067.1 AGBW02011630 OWR46390.1 NWSH01000400 PCG76680.1 NNAY01000053 OXU31504.1 KQ977780 KYM99950.1 GL438292 EFN69190.1 GEDC01020678 GEDC01012952 JAS16620.1 JAS24346.1 GL888262 EGI63689.1 GEDC01029021 GEDC01015563 GEDC01005018 JAS08277.1 JAS21735.1 JAS32280.1 KQ976441 KYM86393.1 KQ983001 KYQ48479.1 ADTU01003040 ADTU01003041 GL766449 EFZ15298.1 KQ979074 KYN22898.1 LBMM01001404 KMQ96380.1 KQ981610 KYN39363.1 KQ435053 KZC14258.1 QOIP01000001 RLU27580.1 GL451029 EFN80176.1 PYGN01000086 PSN54649.1 KZ288258 PBC30512.1 ACPB03015469 ACPB03015470 ACPB03015471 KQ762659 OAD55550.1 KQ435735 KOX77152.1 KQ414574 KOC71253.1 GBYB01005478 JAG75245.1 GBYB01014407 JAG84174.1 GBBI01003159 JAC15553.1 DS235030 EEB10715.1 GEZM01070994 JAV66147.1 GFDF01006855 JAV07229.1 AJWK01006860 AJWK01006861 AJWK01006862 GANO01003076 JAB56795.1 CH477192 EAT48581.1 JXUM01000533 JXUM01000534 AAAB01008804 EAU77730.2 GFDL01011566 JAV23479.1 GFDL01011576 JAV23469.1 AXCN02001143 DS231856 EDS38438.1 GGFJ01000145 MBW49286.1 UFQT01001941 SSX32186.1 GGFM01000903 MBW21654.1 UFQS01000052 UFQT01000052 SSW98661.1 SSX19047.1 ABLF02039321 ABLF02039331 GGFL01000629 MBW64807.1 GGFL01000624 MBW64802.1 GGFL01000628 MBW64806.1 GGFJ01000146 MBW49287.1 GGMR01006057 MBY18676.1 GGFJ01000144 MBW49285.1 ADMH02001385 ETN62687.1 AXCM01006030 GDHF01015414 JAI36900.1 JXJN01007040 JXJN01007041 GGFK01002350 MBW35671.1 CCAG010006850 GGFM01001351 MBW22102.1 GGFK01002343 MBW35664.1 GGFK01002371 MBW35692.1 GAKP01022532 JAC36420.1 GAMC01019691 JAB86864.1 GGFK01003308 MBW36629.1 GGMS01007775 MBY76978.1 GBXI01013809 JAD00483.1 JRES01000610 KNC29902.1 KQ971410 KYB24843.1 ATLV01016620 KE525097 KFB41452.1 KYB24844.1 CH940647 KRF85222.1 EDW70967.1 KRF85218.1 KRF85219.1 KRF85220.1 KRF85221.1 KA645003 AFP59632.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000218220
UP000215335
UP000002358
+ More
UP000078542 UP000000311 UP000007755 UP000078540 UP000075809 UP000005205 UP000078492 UP000036403 UP000078541 UP000076502 UP000279307 UP000008237 UP000245037 UP000005203 UP000242457 UP000015103 UP000053105 UP000053825 UP000009046 UP000092461 UP000008820 UP000069940 UP000007062 UP000075886 UP000002320 UP000069272 UP000007819 UP000075900 UP000075882 UP000000673 UP000075903 UP000075883 UP000092443 UP000092460 UP000092445 UP000092444 UP000078200 UP000075920 UP000075884 UP000076408 UP000075885 UP000075881 UP000091820 UP000095301 UP000095300 UP000037069 UP000007266 UP000030765 UP000075880 UP000008792
UP000078542 UP000000311 UP000007755 UP000078540 UP000075809 UP000005205 UP000078492 UP000036403 UP000078541 UP000076502 UP000279307 UP000008237 UP000245037 UP000005203 UP000242457 UP000015103 UP000053105 UP000053825 UP000009046 UP000092461 UP000008820 UP000069940 UP000007062 UP000075886 UP000002320 UP000069272 UP000007819 UP000075900 UP000075882 UP000000673 UP000075903 UP000075883 UP000092443 UP000092460 UP000092445 UP000092444 UP000078200 UP000075920 UP000075884 UP000076408 UP000075885 UP000075881 UP000091820 UP000095301 UP000095300 UP000037069 UP000007266 UP000030765 UP000075880 UP000008792
PRIDE
Pfam
Interpro
IPR036940
PI3/4_kinase_cat_sf
+ More
IPR001263 PI3K_accessory_dom
IPR036871 PX_dom_sf
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR015433 PI_Kinase
IPR002420 PI3K_C2_dom
IPR042236 PI3K_accessory_sf
IPR000341 PI3K_Ras-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR001683 Phox
IPR000008 C2_dom
IPR002402 Cyt_P450_E_grp-II
IPR036396 Cyt_P450_sf
IPR001128 Cyt_P450
IPR001263 PI3K_accessory_dom
IPR036871 PX_dom_sf
IPR016024 ARM-type_fold
IPR035892 C2_domain_sf
IPR015433 PI_Kinase
IPR002420 PI3K_C2_dom
IPR042236 PI3K_accessory_sf
IPR000341 PI3K_Ras-bd_dom
IPR029071 Ubiquitin-like_domsf
IPR018936 PI3/4_kinase_CS
IPR000403 PI3/4_kinase_cat_dom
IPR011009 Kinase-like_dom_sf
IPR001683 Phox
IPR000008 C2_dom
IPR002402 Cyt_P450_E_grp-II
IPR036396 Cyt_P450_sf
IPR001128 Cyt_P450
SUPFAM
ProteinModelPortal
H9IVI8
A0A2W1BRJ4
A0A194PJA3
A0A2H1VGX9
A0A212EY28
A0A2A4JZ40
+ More
A0A232FL80 K7IW81 A0A151IFN4 E2ABJ5 A0A1B6CT28 F4WQ66 A0A1B6D833 A0A195BML6 A0A151WKS4 A0A158NXE4 E9IVV7 A0A195ECF4 A0A0J7NVG0 A0A195FG44 A0A154PSM0 A0A3L8E4C0 E2BVU7 A0A2P8ZDS3 A0A088AC41 A0A2A3EGV0 T1HF92 A0A310SA10 A0A0N0U690 A0A0L7RKB3 A0A0C9PVJ8 A0A0C9RM03 A0A023F2W8 E0VBF9 A0A1Y1L4Z2 A0A1L8DL86 A0A1B0CCJ7 W4VRG7 Q0IGF6 A0A182GNH9 A0NBA9 A0A1Q3F7H7 A0A1Q3F7G5 A0A182QRR1 B0W7X6 A0A182FTM6 A0A2M4B8C5 A0A336MPB8 A0A2M3YZF9 A0A336LRK7 J9JP72 A0A2M4CHI7 A0A2M4CHG7 A0A2M4CHI5 A0A2M4B8D1 A0A182RUL2 A0A2S2NPJ6 A0A182KKM4 A0A2M4B8A2 W5JEA5 A0A182V420 A0A182MQA5 A0A1A9YEL9 A0A0K8VEC8 A0A1B0B1B1 A0A1B0A2K1 A0A2M4A4J6 A0A1B0FDB1 A0A2M3Z0P3 A0A2M4A4C5 A0A2M4A4P5 A0A1A9VMZ6 A0A034V3S2 A0A182WDV0 A0A182N318 W8BAJ7 A0A182YBJ8 A0A182PSA9 A0A2M4A771 A0A2S2QGW6 A0A0A1WPZ0 A0A182JVT2 A0A1A9X0C7 A0A1I8M8C7 A0A1I8NM56 A0A0L0CCG5 A0A139WA97 A0A084VU08 A0A139WAA5 A0A182IQY2 A0A0Q9WM38 B4LCS5 T1PAR1
A0A232FL80 K7IW81 A0A151IFN4 E2ABJ5 A0A1B6CT28 F4WQ66 A0A1B6D833 A0A195BML6 A0A151WKS4 A0A158NXE4 E9IVV7 A0A195ECF4 A0A0J7NVG0 A0A195FG44 A0A154PSM0 A0A3L8E4C0 E2BVU7 A0A2P8ZDS3 A0A088AC41 A0A2A3EGV0 T1HF92 A0A310SA10 A0A0N0U690 A0A0L7RKB3 A0A0C9PVJ8 A0A0C9RM03 A0A023F2W8 E0VBF9 A0A1Y1L4Z2 A0A1L8DL86 A0A1B0CCJ7 W4VRG7 Q0IGF6 A0A182GNH9 A0NBA9 A0A1Q3F7H7 A0A1Q3F7G5 A0A182QRR1 B0W7X6 A0A182FTM6 A0A2M4B8C5 A0A336MPB8 A0A2M3YZF9 A0A336LRK7 J9JP72 A0A2M4CHI7 A0A2M4CHG7 A0A2M4CHI5 A0A2M4B8D1 A0A182RUL2 A0A2S2NPJ6 A0A182KKM4 A0A2M4B8A2 W5JEA5 A0A182V420 A0A182MQA5 A0A1A9YEL9 A0A0K8VEC8 A0A1B0B1B1 A0A1B0A2K1 A0A2M4A4J6 A0A1B0FDB1 A0A2M3Z0P3 A0A2M4A4C5 A0A2M4A4P5 A0A1A9VMZ6 A0A034V3S2 A0A182WDV0 A0A182N318 W8BAJ7 A0A182YBJ8 A0A182PSA9 A0A2M4A771 A0A2S2QGW6 A0A0A1WPZ0 A0A182JVT2 A0A1A9X0C7 A0A1I8M8C7 A0A1I8NM56 A0A0L0CCG5 A0A139WA97 A0A084VU08 A0A139WAA5 A0A182IQY2 A0A0Q9WM38 B4LCS5 T1PAR1
PDB
4A55
E-value=1.09809e-52,
Score=528
Ontologies
PATHWAY
GO
GO:0048015
GO:0016301
GO:0046854
GO:0035091
GO:0016021
GO:0020037
GO:0016705
GO:0005506
GO:0005524
GO:0004497
GO:0035005
GO:0036092
GO:0005737
GO:0016477
GO:0016303
GO:0005886
GO:0016020
GO:0014065
GO:0005942
GO:0017137
GO:0017124
GO:0042059
GO:0030866
GO:0010008
GO:0005515
GO:0005488
GO:0006508
GO:0016573
GO:0008270
GO:0022891
GO:0003707
GO:0051537
PANTHER
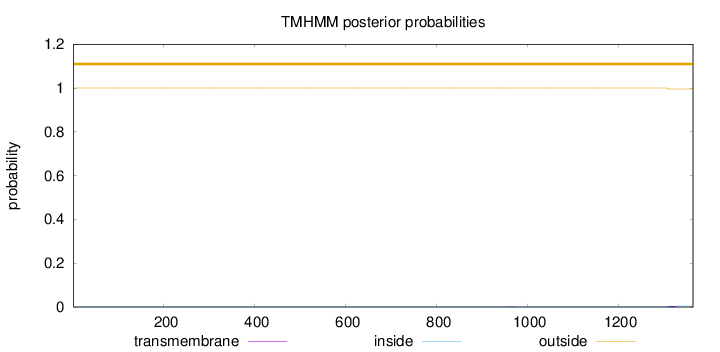
Topology
Length:
1364
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09305
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00001
outside
1 - 1364
Population Genetic Test Statistics
Pi
217.417029
Theta
159.626552
Tajima's D
0.852472
CLR
0.425479
CSRT
0.623868806559672
Interpretation
Uncertain
