Gene
KWMTBOMO07711 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000875
Annotation
carboxyl/choline_esterase_CCE006d_[Helicoverpa_armigera]
Full name
Carboxylic ester hydrolase
Location in the cell
PlasmaMembrane Reliability : 1.322
Sequence
CDS
ATGGATTTCATGCCGTGGTTCTTGTTGCTGATGTTAAGTTACGTTCAGGCTAATCTAGTGAGGGTGGATCCATTAGTTCTTATCAGTAACCAAGGACTAGTGAGGGGTCACAAAGCAGCAGACGGAGATTATTCCATATTTTTGGGTATTCCTTACGCCAAAGTGGACGTAAATAATCCTTTTAGTACTGCTACAGACCCTATAGCTTTTCCTGAAATAATTCACGATGCCGTTGACGAAGTGAGGTGCCCTCAAACATCTTCGTCGGTCGGTGAACAAACACTAAATTGTCTACGTTTGAACATTTACGTACCAAGTGCAGCTAGTTCTAGGAACCCTATGCCTGTCTTAGTTTGGATTCATGGAGGAGATTTTGCGACAGGAAGCGCTTCGGATTACGGAGTCAAAAATATAGTTCGGCATGGAGTTTTAGTAGTTACAATGAATTATAGATTGGGGCCATATGGATTTTTGTGTCTTGATGTTCCAACTGCGTCTGGAAATCAGGGACTTCGGGATCAGTACAAAGCTTTAAGCTGGATTCGAAATAACATTGCGTCTTTTGGAGGCAATCCGTATAATGTAACTATTGCAGGTCAAGATGCCGGAGCCACATCTGTGCTGCTACAGCTGTATTCTGACAACGAAAAACTATTTCACAAGGTTATCATTGAAAGCGGTACACCACAAAGTGAAGGCATGTTCGTAAACGCAGATGTTGAGGCAGCCATTAAATTAGCTGTTCATTTAGGATTGAATACAACTGAAACTGAAGAAGCTATACAATTCCTCACAAAAACATCACCTGACTTAGTAGTTGCTGCTGCTAAAGAATTAGAATTACAATTGAAGCCTTGTAGAGAGAGATCGTTTAGCGGAATCAGCAATTTTGTTGAAAGTGATCCCTATTCTTTGACGAACAAGAAAAAAGTTAGAAACACTCCAATTATGATCGGGAATACAAATAAAGAGAAAAAGTCTTTGAGTGCAGATTACTTTTCAACAGACCCCTTTTATGAAAAGTTAAAGAGTAACTTTAACCTTGACGAAGCTCAAATAGAAAAAGCAGCAAAGAACGTAAGGCATTTTTACATTGGAGACAGCTCTTTATCAGCGGAAGTTGCTTCTGAATTACAAGACTTTGAATCAGACTTTGTTTACAATCACCCGTCTCAAAGAACTATTACACAACTGCTTGACGAAAACGCAGGAAAGATTTACGAGTATTTGTTCAGTTATTCCGGGGAGTTAGAGGATGGCGCTGAGCATTCATCAGAGTTAAACTACTTATTCGAAACGGCTGGAGGTTCAGATAGGACGGAAGAGGAGCAATTAATGGTCGATCGAATAACGACTCTATGGACTAATTTCGTTAAATTCGGGAATCCCACTCCGAAATCGACAGATCTGATTCCTACGATCTGGGAGCCTGTCACTGCCGATACCAGGCCGGTGTTGGTGCTTGACAAGGATGTACGTTTAGACAGCAGGATTGAACGCCAAAGGATGGCGTACTGGGACCTTTTCTTCACAACGTACGGACAATACAATTTTATAGCAAGAGAAACGTGCGTATTCAAAGACTATGATGACTAA
Protein
MDFMPWFLLLMLSYVQANLVRVDPLVLISNQGLVRGHKAADGDYSIFLGIPYAKVDVNNPFSTATDPIAFPEIIHDAVDEVRCPQTSSSVGEQTLNCLRLNIYVPSAASSRNPMPVLVWIHGGDFATGSASDYGVKNIVRHGVLVVTMNYRLGPYGFLCLDVPTASGNQGLRDQYKALSWIRNNIASFGGNPYNVTIAGQDAGATSVLLQLYSDNEKLFHKVIIESGTPQSEGMFVNADVEAAIKLAVHLGLNTTETEEAIQFLTKTSPDLVVAAAKELELQLKPCRERSFSGISNFVESDPYSLTNKKKVRNTPIMIGNTNKEKKSLSADYFSTDPFYEKLKSNFNLDEAQIEKAAKNVRHFYIGDSSLSAEVASELQDFESDFVYNHPSQRTITQLLDENAGKIYEYLFSYSGELEDGAEHSSELNYLFETAGGSDRTEEEQLMVDRITTLWTNFVKFGNPTPKSTDLIPTIWEPVTADTRPVLVLDKDVRLDSRIERQRMAYWDLFFTTYGQYNFIARETCVFKDYDD
Summary
Similarity
Belongs to the type-B carboxylesterase/lipase family.
Feature
chain Carboxylic ester hydrolase
Uniprot
H9IUE5
D5G3E4
A0A2H1VH39
A0A2A4JYV0
A0A2W1BNW5
A0A0L7LKE5
+ More
A0A194QYN7 A0A194PD51 A0A1U9X1U4 A0A0B4RY83 A0A1U9X1X9 A0A1U9X1Z4 A0A212EY53 A0A1L8D6M4 A0A1L8D6W6 A0A1L8D6U2 A0A212FJ60 A0A0N1IPF0 A0A1L8D6H6 A0A194Q254 H9J1Y0 A0A2H1VCT7 A0A2H1W648 A0A194PEF7 A0A2W1BZX4 A0A2I6RJM2 H9J066 A4UA26 A0A2H1VCP0 A0A2H1VCN6 A0A2H1VCQ9 A0A194PJ15 B1Q141 M1RGF2 A0A2A4K7E9 H9IUA7 A0A1J0M1Z7 A0A2H1VCN0 A0A1U9X1V1 A0A194Q2Q8 A0A2H1VAJ0 A0A3G1T174 D3GDM3 A0A194Q887 A0A2A4K075 A0A2H1WRP2 A0A2A4JZ33 A0A2A4JU91 B3GEF6 A0A2W1BXP7 A0A0B4RXS5 H9IZW1 D9ILE0 A0A2A4JSH8 A0A1U9X1V6 A0A2A4JR93 A0A0C5C1P7 A0A194Q249 A0A0F7QHL9 A0A0N1PJ90 A0A194PDI7 D5G3E3 A0A2A4JAV9 A0A0N1IGW1 A0A3S2LSF5 H9J067 A0A212FJ67 A0A291P0Q2 A0A291P105 A0A2W1BJ27 D5G3E0 L7P6C6 A0A1U9X1U7 A0A2W1BQY3 A0A076FR85 B1Q137 A0A194RIH0 A0A2H1VJA9 A0A2A4JU71 A0A2W1BK86 A0A0K0XRM2 A0A2W1BTR6 D5G3E2 D5G3E1 D3GDL7 A0A0L7L0K6 A0A1U9X1V3 A0A2W1BTK5 A0A2H1VCR3 A0A1U9X1V0 A0A2H1VEA4 H9IWJ7 A0A2W1BVP4 A0A2H1VAI9 A0A0U4D5F7 A0A0K0XRP4 A0A0L7L3V0 A0A1U9X1Z0 A0A0L7LED6 A0A0L7L1R0
A0A194QYN7 A0A194PD51 A0A1U9X1U4 A0A0B4RY83 A0A1U9X1X9 A0A1U9X1Z4 A0A212EY53 A0A1L8D6M4 A0A1L8D6W6 A0A1L8D6U2 A0A212FJ60 A0A0N1IPF0 A0A1L8D6H6 A0A194Q254 H9J1Y0 A0A2H1VCT7 A0A2H1W648 A0A194PEF7 A0A2W1BZX4 A0A2I6RJM2 H9J066 A4UA26 A0A2H1VCP0 A0A2H1VCN6 A0A2H1VCQ9 A0A194PJ15 B1Q141 M1RGF2 A0A2A4K7E9 H9IUA7 A0A1J0M1Z7 A0A2H1VCN0 A0A1U9X1V1 A0A194Q2Q8 A0A2H1VAJ0 A0A3G1T174 D3GDM3 A0A194Q887 A0A2A4K075 A0A2H1WRP2 A0A2A4JZ33 A0A2A4JU91 B3GEF6 A0A2W1BXP7 A0A0B4RXS5 H9IZW1 D9ILE0 A0A2A4JSH8 A0A1U9X1V6 A0A2A4JR93 A0A0C5C1P7 A0A194Q249 A0A0F7QHL9 A0A0N1PJ90 A0A194PDI7 D5G3E3 A0A2A4JAV9 A0A0N1IGW1 A0A3S2LSF5 H9J067 A0A212FJ67 A0A291P0Q2 A0A291P105 A0A2W1BJ27 D5G3E0 L7P6C6 A0A1U9X1U7 A0A2W1BQY3 A0A076FR85 B1Q137 A0A194RIH0 A0A2H1VJA9 A0A2A4JU71 A0A2W1BK86 A0A0K0XRM2 A0A2W1BTR6 D5G3E2 D5G3E1 D3GDL7 A0A0L7L0K6 A0A1U9X1V3 A0A2W1BTK5 A0A2H1VCR3 A0A1U9X1V0 A0A2H1VEA4 H9IWJ7 A0A2W1BVP4 A0A2H1VAI9 A0A0U4D5F7 A0A0K0XRP4 A0A0L7L3V0 A0A1U9X1Z0 A0A0L7LED6 A0A0L7L1R0
EC Number
3.1.1.-
Pubmed
EMBL
BABH01001182
KC200271
AGG20206.1
FJ997308
ADF43473.1
ODYU01002500
+ More
SOQ40071.1 NWSH01000400 PCG76673.1 KZ149946 PZC76749.1 JTDY01000777 KOB75922.1 KQ460947 KPJ10567.1 KQ459606 KPI91162.1 KY021838 AQY62727.1 KJ023211 AIY69039.1 KY021873 AQY62762.1 KY021884 AQY62773.1 AGBW02011630 OWR46387.1 GEYN01000027 JAV02102.1 GEYN01000045 JAV02084.1 GEYN01000065 JAV02064.1 AGBW02008309 OWR53771.1 KQ460400 KPJ14974.1 GEYN01000070 JAV02059.1 KQ459564 KPI99621.1 BABH01007753 ODYU01001845 SOQ38611.1 SOQ38612.1 ODYU01006598 SOQ48575.1 KPI91074.1 KZ149903 PZC78410.1 KY304479 AUN86410.1 BABH01018991 BABH01018992 DQ680829 ABH01082.1 SOQ38610.1 SOQ38609.1 SOQ38607.1 KPI91075.1 EU523536 ACB12415.1 KC200270 AGG20205.1 NWSH01000067 PCG79999.1 BABH01001500 KU375432 APD13874.1 SOQ38608.1 KY021842 AQY62731.1 KPI99618.1 ODYU01001525 SOQ37859.1 MG846884 AXY94736.1 FJ652455 ACV60239.1 KPI99620.1 NWSH01000356 PCG77070.1 ODYU01010533 SOQ55708.1 PCG77069.1 NWSH01000660 PCG74962.1 EU688968 ACD80422.1 PZC78414.1 KJ023212 AIY69040.1 BABH01019018 HM150791 ADI47117.1 PCG74961.1 KY021858 AQY62747.1 NWSH01000834 PCG73942.1 KP001149 AJN91194.1 KPI99616.1 LC017770 BAR64784.1 KQ459837 KPJ19757.1 KPI91073.1 FJ997307 ADF43472.1 NWSH01002285 PCG68674.1 KPJ19760.1 RSAL01000258 RVE43396.1 BABH01018995 OWR53772.1 MF687561 ATJ44487.1 MF687628 ATJ44554.1 KZ150117 PZC73247.1 FJ997304 ADF43469.1 JX306730 AFO65061.1 KY021848 AQY62737.1 KZ150007 PZC75160.1 KF960780 AII21982.1 EU523532 ACB12411.1 KQ460205 KPJ17125.1 ODYU01002878 SOQ40908.1 PCG74959.1 PCG74960.1 PZC73246.1 KP938883 AKS40364.1 PZC78409.1 FJ997306 ADF43471.1 FJ997305 ADF43470.1 FJ652449 ACV60233.1 JTDY01003780 KOB69017.1 KY021841 AQY62730.1 PZC78412.1 SOQ38605.1 KY021850 AQY62739.1 SOQ38604.1 BABH01021273 PZC78411.1 SOQ37858.1 KU175691 ALX37953.1 KP938882 AKS40363.1 JTDY01003106 KOB70142.1 KY021887 AQY62776.1 JTDY01001549 KOB73566.1 JTDY01003509 KOB69443.1
SOQ40071.1 NWSH01000400 PCG76673.1 KZ149946 PZC76749.1 JTDY01000777 KOB75922.1 KQ460947 KPJ10567.1 KQ459606 KPI91162.1 KY021838 AQY62727.1 KJ023211 AIY69039.1 KY021873 AQY62762.1 KY021884 AQY62773.1 AGBW02011630 OWR46387.1 GEYN01000027 JAV02102.1 GEYN01000045 JAV02084.1 GEYN01000065 JAV02064.1 AGBW02008309 OWR53771.1 KQ460400 KPJ14974.1 GEYN01000070 JAV02059.1 KQ459564 KPI99621.1 BABH01007753 ODYU01001845 SOQ38611.1 SOQ38612.1 ODYU01006598 SOQ48575.1 KPI91074.1 KZ149903 PZC78410.1 KY304479 AUN86410.1 BABH01018991 BABH01018992 DQ680829 ABH01082.1 SOQ38610.1 SOQ38609.1 SOQ38607.1 KPI91075.1 EU523536 ACB12415.1 KC200270 AGG20205.1 NWSH01000067 PCG79999.1 BABH01001500 KU375432 APD13874.1 SOQ38608.1 KY021842 AQY62731.1 KPI99618.1 ODYU01001525 SOQ37859.1 MG846884 AXY94736.1 FJ652455 ACV60239.1 KPI99620.1 NWSH01000356 PCG77070.1 ODYU01010533 SOQ55708.1 PCG77069.1 NWSH01000660 PCG74962.1 EU688968 ACD80422.1 PZC78414.1 KJ023212 AIY69040.1 BABH01019018 HM150791 ADI47117.1 PCG74961.1 KY021858 AQY62747.1 NWSH01000834 PCG73942.1 KP001149 AJN91194.1 KPI99616.1 LC017770 BAR64784.1 KQ459837 KPJ19757.1 KPI91073.1 FJ997307 ADF43472.1 NWSH01002285 PCG68674.1 KPJ19760.1 RSAL01000258 RVE43396.1 BABH01018995 OWR53772.1 MF687561 ATJ44487.1 MF687628 ATJ44554.1 KZ150117 PZC73247.1 FJ997304 ADF43469.1 JX306730 AFO65061.1 KY021848 AQY62737.1 KZ150007 PZC75160.1 KF960780 AII21982.1 EU523532 ACB12411.1 KQ460205 KPJ17125.1 ODYU01002878 SOQ40908.1 PCG74959.1 PCG74960.1 PZC73246.1 KP938883 AKS40364.1 PZC78409.1 FJ997306 ADF43471.1 FJ997305 ADF43470.1 FJ652449 ACV60233.1 JTDY01003780 KOB69017.1 KY021841 AQY62730.1 PZC78412.1 SOQ38605.1 KY021850 AQY62739.1 SOQ38604.1 BABH01021273 PZC78411.1 SOQ37858.1 KU175691 ALX37953.1 KP938882 AKS40363.1 JTDY01003106 KOB70142.1 KY021887 AQY62776.1 JTDY01001549 KOB73566.1 JTDY01003509 KOB69443.1
Proteomes
Pfam
PF00135 COesterase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9IUE5
D5G3E4
A0A2H1VH39
A0A2A4JYV0
A0A2W1BNW5
A0A0L7LKE5
+ More
A0A194QYN7 A0A194PD51 A0A1U9X1U4 A0A0B4RY83 A0A1U9X1X9 A0A1U9X1Z4 A0A212EY53 A0A1L8D6M4 A0A1L8D6W6 A0A1L8D6U2 A0A212FJ60 A0A0N1IPF0 A0A1L8D6H6 A0A194Q254 H9J1Y0 A0A2H1VCT7 A0A2H1W648 A0A194PEF7 A0A2W1BZX4 A0A2I6RJM2 H9J066 A4UA26 A0A2H1VCP0 A0A2H1VCN6 A0A2H1VCQ9 A0A194PJ15 B1Q141 M1RGF2 A0A2A4K7E9 H9IUA7 A0A1J0M1Z7 A0A2H1VCN0 A0A1U9X1V1 A0A194Q2Q8 A0A2H1VAJ0 A0A3G1T174 D3GDM3 A0A194Q887 A0A2A4K075 A0A2H1WRP2 A0A2A4JZ33 A0A2A4JU91 B3GEF6 A0A2W1BXP7 A0A0B4RXS5 H9IZW1 D9ILE0 A0A2A4JSH8 A0A1U9X1V6 A0A2A4JR93 A0A0C5C1P7 A0A194Q249 A0A0F7QHL9 A0A0N1PJ90 A0A194PDI7 D5G3E3 A0A2A4JAV9 A0A0N1IGW1 A0A3S2LSF5 H9J067 A0A212FJ67 A0A291P0Q2 A0A291P105 A0A2W1BJ27 D5G3E0 L7P6C6 A0A1U9X1U7 A0A2W1BQY3 A0A076FR85 B1Q137 A0A194RIH0 A0A2H1VJA9 A0A2A4JU71 A0A2W1BK86 A0A0K0XRM2 A0A2W1BTR6 D5G3E2 D5G3E1 D3GDL7 A0A0L7L0K6 A0A1U9X1V3 A0A2W1BTK5 A0A2H1VCR3 A0A1U9X1V0 A0A2H1VEA4 H9IWJ7 A0A2W1BVP4 A0A2H1VAI9 A0A0U4D5F7 A0A0K0XRP4 A0A0L7L3V0 A0A1U9X1Z0 A0A0L7LED6 A0A0L7L1R0
A0A194QYN7 A0A194PD51 A0A1U9X1U4 A0A0B4RY83 A0A1U9X1X9 A0A1U9X1Z4 A0A212EY53 A0A1L8D6M4 A0A1L8D6W6 A0A1L8D6U2 A0A212FJ60 A0A0N1IPF0 A0A1L8D6H6 A0A194Q254 H9J1Y0 A0A2H1VCT7 A0A2H1W648 A0A194PEF7 A0A2W1BZX4 A0A2I6RJM2 H9J066 A4UA26 A0A2H1VCP0 A0A2H1VCN6 A0A2H1VCQ9 A0A194PJ15 B1Q141 M1RGF2 A0A2A4K7E9 H9IUA7 A0A1J0M1Z7 A0A2H1VCN0 A0A1U9X1V1 A0A194Q2Q8 A0A2H1VAJ0 A0A3G1T174 D3GDM3 A0A194Q887 A0A2A4K075 A0A2H1WRP2 A0A2A4JZ33 A0A2A4JU91 B3GEF6 A0A2W1BXP7 A0A0B4RXS5 H9IZW1 D9ILE0 A0A2A4JSH8 A0A1U9X1V6 A0A2A4JR93 A0A0C5C1P7 A0A194Q249 A0A0F7QHL9 A0A0N1PJ90 A0A194PDI7 D5G3E3 A0A2A4JAV9 A0A0N1IGW1 A0A3S2LSF5 H9J067 A0A212FJ67 A0A291P0Q2 A0A291P105 A0A2W1BJ27 D5G3E0 L7P6C6 A0A1U9X1U7 A0A2W1BQY3 A0A076FR85 B1Q137 A0A194RIH0 A0A2H1VJA9 A0A2A4JU71 A0A2W1BK86 A0A0K0XRM2 A0A2W1BTR6 D5G3E2 D5G3E1 D3GDL7 A0A0L7L0K6 A0A1U9X1V3 A0A2W1BTK5 A0A2H1VCR3 A0A1U9X1V0 A0A2H1VEA4 H9IWJ7 A0A2W1BVP4 A0A2H1VAI9 A0A0U4D5F7 A0A0K0XRP4 A0A0L7L3V0 A0A1U9X1Z0 A0A0L7LED6 A0A0L7L1R0
PDB
2XUQ
E-value=3.86884e-40,
Score=415
Ontologies
Topology
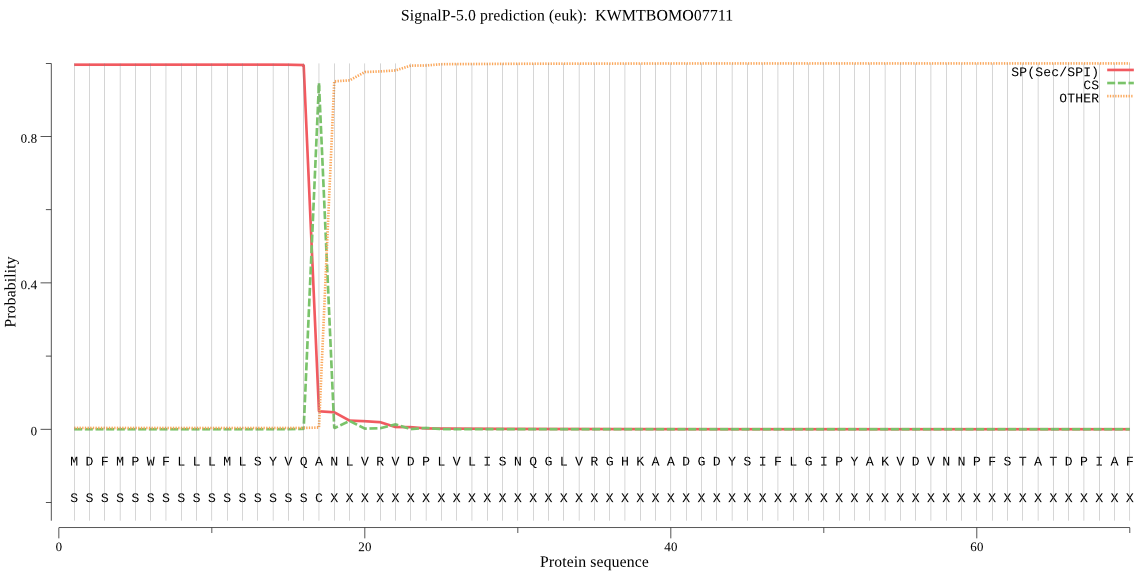
SignalP
Position: 1 - 17,
Likelihood: 0.995915
Length:
531
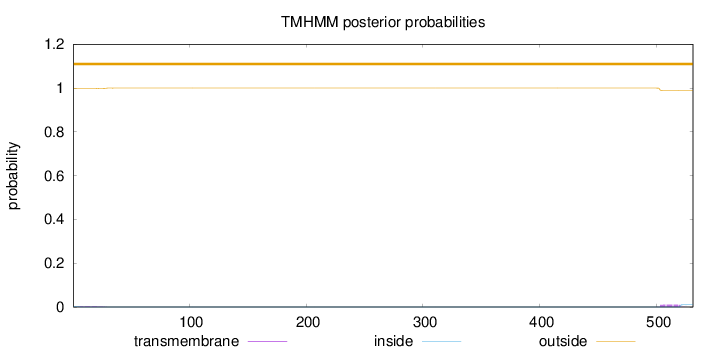
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25571
Exp number, first 60 AAs:
0.06441
Total prob of N-in:
0.00316
outside
1 - 531
Population Genetic Test Statistics
Pi
228.315096
Theta
200.746398
Tajima's D
0.252113
CLR
0.325508
CSRT
0.438878056097195
Interpretation
Uncertain
