Gene
KWMTBOMO07701 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA001272
Annotation
PREDICTED:_venom_dipeptidyl_peptidase_4-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.092 PlasmaMembrane Reliability : 1.505
Sequence
CDS
ATGCACCGAACTCACAAGAGGCCCTACCACCATATTCCGAGAATCACCGCTCTTGCTAGGGCTGGTGTTACCAAATTCTCGCAGCTGTTCAGATACAGTTTCTTGGCTCGCTACGCTGCTTATGACGTCGCGACCGGCTCTGAACGAGACATCGTCCCTCCTGGAGTACAATCCAGCGAGGTCTTCCTCCAGAACTTCGTGTGGGGTCCCAGAGGCACATCTCTCGCTTTCGTCTATTTGAACAACATTTTCTATCAGTCGAGCTTGACGGCCGCCCCTCAACAGATTACGACTACAGGCGAGATTAATGTCATCTATCACGGTGTTCCCGATTGGGTCTATGAAGAGGAAGTCTTCAGTTCAAACAACGCGATGTGGTTCTCGGCGGATGGTCAGAAGTTAGCCTACGTGACTTTTGACGATACAAACGTTCGGGTGATGCCGGTCCCGCATTACGGCGTCCCAGGCAACATCAACTACCAATACACCCAGCACCATCAAATAAGATATCCTAAGCCCGGCACAACCAATCCAACTGTTACGGTTACTCTGCGGGATCTCAATTCTGGCTCTTCGAATGCGTACGCTGCCCCCAGCGATCTTAACGAGCCGATCTTGAGGACAGTCCAGTTCACAGCTAACAACGTGATCGCCTTGATGTGGACTAACCGAGTGCAGACAACCCTAAGGGTTGTGCTATGCACTCTCGGACAAACGGCTTGTACACAGATATACCAATACACAGAAACTAACGGATGGATCGACAATATTCCACTGCTCTTTAATAACGCGGGCGATTCTTTCGTGACGATTTTGCCTTATGCTGTAGATGGAGTTCGATACAAGCAGATTGTGCAAGTTTCACGAGGCACTGGCACAACTTGGCAGAGTACGGGAAGGACCAACACAGCTCACACCGTTTCGGAAATCCTGCACTGGTCTCCGAATGATATCATCTGGTACCGAGCCACACGGGTTAACGACCCCGCAGAACAACATATATACACCGTGAACAGCGCCCGTGAGGTTGCGTGCTTCACTTGCGAGATCCGAAGGCTGGACGGCGGACAGTGCTTGTATAACGAAGCTACCATCAGTTCGAGTGGCGAGCGGATCACGATCAACTGTGCCGGACCTGACGTTCCGCAAGTTCTCATCTTCGACAGTAGTGGTAACCTTATCAAAGAATGGGACACAAACTCGGAGCTCTTATCGTTGGTGGCCAACAAAATTCTTCCGACCACAGTGCGGATGTCCGTACCAGTTGCCGAAGGGTATCCCAACGCTGATGTCCAGATACAAGTTCCAGCAGATTACCAGCACAGGACCAATTTGCCTCTTCTTGTTTATGTGTACGGCGGGCCCGACACATCACTGGTAACGAAGCAGTGGTCGATTGACTGGGGTAGCTACCTTGTCAGTCGCTGGAACGTCGCCGTGGCTCATATCGACGGTCGTGGAGCTGGAAACCGAGGCGTTAACAATTTGTTTGCTCTTAACAGAAAACTTGGTACCGTGGAGATAGAAGATCAGATTTCTGTTACAAAGTATTTACAAGAAAAACTTTCGTATGTCGATGCTAATCGCACCTGTATCTGGGGCTGGTCGTACGGCGGCTACGCGGCCTCCCTGGCTCTCGCCCGTGGTGGAGATGTATTCCGATGTGCTGCTGCAGTCGCTCCTGTTGTCGACTGGAAATTCTATGATACAATATACACGGAACGTTACATGGATCTGCCACAGAACAACGTAGAGGGCTACGCCGCTTCATCGCTGTTGGTCAATGATGTGGTGGATTCATACAAAAGTAAAAGCTATTTCCTGGTTCACGGCACAGCGGACGACAATGTGCATTACCAACACGCTATGCTGATCTCGCGGCTTTTGCAGAGGCGGGACATCTACTTCACACAAATGAGCTACACCGATGAAGACCACGATCTAATTGGCGTGAGGCCGCATCTTTACCACGGCCTAGAGAAGTTCCTTCGAGAAAACATGTTCTAA
Protein
MHRTHKRPYHHIPRITALARAGVTKFSQLFRYSFLARYAAYDVATGSERDIVPPGVQSSEVFLQNFVWGPRGTSLAFVYLNNIFYQSSLTAAPQQITTTGEINVIYHGVPDWVYEEEVFSSNNAMWFSADGQKLAYVTFDDTNVRVMPVPHYGVPGNINYQYTQHHQIRYPKPGTTNPTVTVTLRDLNSGSSNAYAAPSDLNEPILRTVQFTANNVIALMWTNRVQTTLRVVLCTLGQTACTQIYQYTETNGWIDNIPLLFNNAGDSFVTILPYAVDGVRYKQIVQVSRGTGTTWQSTGRTNTAHTVSEILHWSPNDIIWYRATRVNDPAEQHIYTVNSAREVACFTCEIRRLDGGQCLYNEATISSSGERITINCAGPDVPQVLIFDSSGNLIKEWDTNSELLSLVANKILPTTVRMSVPVAEGYPNADVQIQVPADYQHRTNLPLLVYVYGGPDTSLVTKQWSIDWGSYLVSRWNVAVAHIDGRGAGNRGVNNLFALNRKLGTVEIEDQISVTKYLQEKLSYVDANRTCIWGWSYGGYAASLALARGGDVFRCAAAVAPVVDWKFYDTIYTERYMDLPQNNVEGYAASSLLVNDVVDSYKSKSYFLVHGTADDNVHYQHAMLISRLLQRRDIYFTQMSYTDEDHDLIGVRPHLYHGLEKFLRENMF
Summary
Similarity
Belongs to the peptidase S9B family.
Uniprot
H9IVJ2
A0A194PEN4
A0A2W1BVE6
A0A2A4JGT3
A0A194QYE1
A0A212EZB6
+ More
A0A3S2LKJ5 A0A0L7LB88 A0A2H1WTE2 A0A1W4X609 A0A1W4X5V5 A0A1W4X5V0 E0VBV5 K7IXE7 E2C5M6 A0A1Y1KXZ8 A0A348G604 A0A158NN55 A0A0P4W4J7 T1I808 A0A146KL11 A0A0A9XS95 A0A0K8SCK7 A0A069DX38 A0A023EZT7 A0A0V0G4N0 A0A0C9RFV4 A0A0C9Q8K2 A0A0L7R316 A0A195C295 A0A1L8E265 A0A1L8E290 A0A0L0CEC5 A0A182H1M1 A0A1B0CY21 Q16RL5 T1DG95 A0A1L8DTH7 A0A0K2UJ39 U5EWK1 A0A232FMA1 A0A2A3E6J8 A0A336K9H3 A0A1J1J458 A0A084WGL0 A0A087ZUP7 A0A1B6DL67 A0A2J7Q7T3 A0A1B0AVV4 A0A1B6EG93 A0A1A9VFE3 A0A1B0G9Y4 A0A1A9XR98 B0WYN8 A0A2M4BE23 A0A182FBI4 A0A2M4BE95 W5J3Y4 A0A1I8MPC7 A0A0R3P7Z3 A0A2M4BDC8 A0A0R3P7F9 A0A182JLE1 A0A1I8MPC6 A0A182RAP7 A0A1A9WD61 A0A182MBV6 M9PCH6 A0A2M4CIB2 A0A1W4U9C0 Q9VUH1 A0A2M3ZFZ0 A0A0Q5U713 A0A2M4A0T9 A0A2M4A0X2 A0A2M3YZQ0 A0A0R1E3Q4 A0A182Q376 A0A2M3YZQ4 A0A182Y621 A0A2M4A0V0 A0A0R3P300 A0A182LP15 A0A182URD4 A0A3R7LWT5 A0A1S4H707 Q7PSF9 A0A182UKA9 B4LF51 A0A182HN75 A0A1W4UKP7 A0A3B0J3T2 A0A0Q5UB85 Q29R16 A0A182X7R6 A0A182WEQ8 A0A182PUL1 A0A0R1DY35 A0A0A1X6T7 A0A0J9RVJ9
A0A3S2LKJ5 A0A0L7LB88 A0A2H1WTE2 A0A1W4X609 A0A1W4X5V5 A0A1W4X5V0 E0VBV5 K7IXE7 E2C5M6 A0A1Y1KXZ8 A0A348G604 A0A158NN55 A0A0P4W4J7 T1I808 A0A146KL11 A0A0A9XS95 A0A0K8SCK7 A0A069DX38 A0A023EZT7 A0A0V0G4N0 A0A0C9RFV4 A0A0C9Q8K2 A0A0L7R316 A0A195C295 A0A1L8E265 A0A1L8E290 A0A0L0CEC5 A0A182H1M1 A0A1B0CY21 Q16RL5 T1DG95 A0A1L8DTH7 A0A0K2UJ39 U5EWK1 A0A232FMA1 A0A2A3E6J8 A0A336K9H3 A0A1J1J458 A0A084WGL0 A0A087ZUP7 A0A1B6DL67 A0A2J7Q7T3 A0A1B0AVV4 A0A1B6EG93 A0A1A9VFE3 A0A1B0G9Y4 A0A1A9XR98 B0WYN8 A0A2M4BE23 A0A182FBI4 A0A2M4BE95 W5J3Y4 A0A1I8MPC7 A0A0R3P7Z3 A0A2M4BDC8 A0A0R3P7F9 A0A182JLE1 A0A1I8MPC6 A0A182RAP7 A0A1A9WD61 A0A182MBV6 M9PCH6 A0A2M4CIB2 A0A1W4U9C0 Q9VUH1 A0A2M3ZFZ0 A0A0Q5U713 A0A2M4A0T9 A0A2M4A0X2 A0A2M3YZQ0 A0A0R1E3Q4 A0A182Q376 A0A2M3YZQ4 A0A182Y621 A0A2M4A0V0 A0A0R3P300 A0A182LP15 A0A182URD4 A0A3R7LWT5 A0A1S4H707 Q7PSF9 A0A182UKA9 B4LF51 A0A182HN75 A0A1W4UKP7 A0A3B0J3T2 A0A0Q5UB85 Q29R16 A0A182X7R6 A0A182WEQ8 A0A182PUL1 A0A0R1DY35 A0A0A1X6T7 A0A0J9RVJ9
Pubmed
19121390
26354079
28756777
22118469
26227816
20566863
+ More
20075255 20798317 28004739 21347285 27129103 26823975 25401762 26334808 25474469 26108605 26483478 17510324 24330624 28648823 24438588 20920257 23761445 25315136 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20966253 12364791 18057021 25830018 22936249
20075255 20798317 28004739 21347285 27129103 26823975 25401762 26334808 25474469 26108605 26483478 17510324 24330624 28648823 24438588 20920257 23761445 25315136 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25244985 20966253 12364791 18057021 25830018 22936249
EMBL
BABH01001205
BABH01001206
BABH01001207
BABH01001208
KQ459606
KPI91154.1
+ More
KZ149946 PZC76760.1 NWSH01001634 PCG70630.1 KQ460947 KPJ10558.1 AGBW02011306 OWR46845.1 RSAL01000080 RVE48668.1 JTDY01001875 KOB72660.1 ODYU01010917 SOQ56323.1 DS235042 EEB10861.1 AAZX01000008 AAZX01000010 AAZX01000041 AAZX01005042 AAZX01009452 AAZX01009962 AAZX01013677 AAZX01015067 AAZX01019562 AAZX01025043 GL452776 EFN76737.1 GEZM01070829 JAV66262.1 FX985543 BBF97877.1 ADTU01020861 ADTU01020862 ADTU01020863 ADTU01020864 ADTU01020865 ADTU01020866 ADTU01020867 ADTU01020868 ADTU01020869 ADTU01020870 GDKW01000335 JAI56260.1 ACPB03004689 GDHC01021358 JAP97270.1 GBHO01021085 GBHO01021084 GBHO01021083 GBHO01015182 JAG22519.1 JAG22520.1 JAG22521.1 JAG28422.1 GBRD01015321 JAG50505.1 GBGD01000523 JAC88366.1 GBBI01003949 JAC14763.1 GECL01003131 JAP02993.1 GBYB01007235 GBYB01010567 JAG77002.1 JAG80334.1 GBYB01010568 JAG80335.1 KQ414663 KOC65219.1 KQ978344 KYM94984.1 GFDF01001254 JAV12830.1 GFDF01001253 JAV12831.1 JRES01000502 KNC30605.1 JXUM01103618 KQ564819 KXJ71707.1 AJWK01035258 AJWK01035259 CH477704 EAT37053.1 GALA01001766 JAA93086.1 GFDF01004363 JAV09721.1 HACA01020704 CDW38065.1 GANO01000531 JAB59340.1 NNAY01000020 OXU31876.1 KZ288351 PBC27345.1 UFQS01000204 UFQT01000204 SSX01299.1 SSX21679.1 CVRI01000070 CRL07192.1 ATLV01023598 ATLV01023599 ATLV01023600 KE525345 KFB49354.1 GEDC01010864 JAS26434.1 NEVH01017440 PNF24641.1 JXJN01004399 GEDC01000404 JAS36894.1 CCAG010008278 DS232195 EDS37148.1 GGFJ01002151 MBW51292.1 GGFJ01002221 MBW51362.1 ADMH02002103 ETN59092.1 CH379069 KRT07558.1 GGFJ01001904 MBW51045.1 KRT07557.1 AXCM01009408 AE014296 AGB94537.1 GGFL01000797 MBW64975.1 AY058514 AAF49712.1 AAL13743.1 GGFM01006617 MBW27368.1 CH954178 KQS43964.1 GGFK01001058 MBW34379.1 GGFK01001049 MBW34370.1 GGFM01001008 MBW21759.1 CM000159 KRK01895.1 AXCN02001048 GGFM01000999 MBW21750.1 GGFK01001051 MBW34372.1 KRT07556.1 QCYY01002917 ROT66607.1 AAAB01008834 EAA05700.5 CH940647 EDW70239.2 KRF84797.1 KRF84799.1 APCN01001877 OUUW01000002 SPP76057.1 KQS43965.1 KQS43966.1 BT024224 ABC86286.1 ACL83310.1 KRK01893.1 KRK01894.1 GBXI01007832 GBXI01000148 JAD06460.1 JAD14144.1 CM002912 KMY99602.1
KZ149946 PZC76760.1 NWSH01001634 PCG70630.1 KQ460947 KPJ10558.1 AGBW02011306 OWR46845.1 RSAL01000080 RVE48668.1 JTDY01001875 KOB72660.1 ODYU01010917 SOQ56323.1 DS235042 EEB10861.1 AAZX01000008 AAZX01000010 AAZX01000041 AAZX01005042 AAZX01009452 AAZX01009962 AAZX01013677 AAZX01015067 AAZX01019562 AAZX01025043 GL452776 EFN76737.1 GEZM01070829 JAV66262.1 FX985543 BBF97877.1 ADTU01020861 ADTU01020862 ADTU01020863 ADTU01020864 ADTU01020865 ADTU01020866 ADTU01020867 ADTU01020868 ADTU01020869 ADTU01020870 GDKW01000335 JAI56260.1 ACPB03004689 GDHC01021358 JAP97270.1 GBHO01021085 GBHO01021084 GBHO01021083 GBHO01015182 JAG22519.1 JAG22520.1 JAG22521.1 JAG28422.1 GBRD01015321 JAG50505.1 GBGD01000523 JAC88366.1 GBBI01003949 JAC14763.1 GECL01003131 JAP02993.1 GBYB01007235 GBYB01010567 JAG77002.1 JAG80334.1 GBYB01010568 JAG80335.1 KQ414663 KOC65219.1 KQ978344 KYM94984.1 GFDF01001254 JAV12830.1 GFDF01001253 JAV12831.1 JRES01000502 KNC30605.1 JXUM01103618 KQ564819 KXJ71707.1 AJWK01035258 AJWK01035259 CH477704 EAT37053.1 GALA01001766 JAA93086.1 GFDF01004363 JAV09721.1 HACA01020704 CDW38065.1 GANO01000531 JAB59340.1 NNAY01000020 OXU31876.1 KZ288351 PBC27345.1 UFQS01000204 UFQT01000204 SSX01299.1 SSX21679.1 CVRI01000070 CRL07192.1 ATLV01023598 ATLV01023599 ATLV01023600 KE525345 KFB49354.1 GEDC01010864 JAS26434.1 NEVH01017440 PNF24641.1 JXJN01004399 GEDC01000404 JAS36894.1 CCAG010008278 DS232195 EDS37148.1 GGFJ01002151 MBW51292.1 GGFJ01002221 MBW51362.1 ADMH02002103 ETN59092.1 CH379069 KRT07558.1 GGFJ01001904 MBW51045.1 KRT07557.1 AXCM01009408 AE014296 AGB94537.1 GGFL01000797 MBW64975.1 AY058514 AAF49712.1 AAL13743.1 GGFM01006617 MBW27368.1 CH954178 KQS43964.1 GGFK01001058 MBW34379.1 GGFK01001049 MBW34370.1 GGFM01001008 MBW21759.1 CM000159 KRK01895.1 AXCN02001048 GGFM01000999 MBW21750.1 GGFK01001051 MBW34372.1 KRT07556.1 QCYY01002917 ROT66607.1 AAAB01008834 EAA05700.5 CH940647 EDW70239.2 KRF84797.1 KRF84799.1 APCN01001877 OUUW01000002 SPP76057.1 KQS43965.1 KQS43966.1 BT024224 ABC86286.1 ACL83310.1 KRK01893.1 KRK01894.1 GBXI01007832 GBXI01000148 JAD06460.1 JAD14144.1 CM002912 KMY99602.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000192223 UP000009046 UP000002358 UP000008237 UP000005205 UP000015103 UP000053825 UP000078542 UP000037069 UP000069940 UP000249989 UP000092461 UP000008820 UP000215335 UP000242457 UP000183832 UP000030765 UP000005203 UP000235965 UP000092460 UP000078200 UP000092444 UP000092443 UP000002320 UP000069272 UP000000673 UP000095301 UP000001819 UP000075880 UP000075900 UP000091820 UP000075883 UP000000803 UP000192221 UP000008711 UP000002282 UP000075886 UP000076408 UP000075882 UP000075903 UP000283509 UP000007062 UP000075902 UP000008792 UP000075840 UP000268350 UP000076407 UP000075920 UP000075885
UP000037510 UP000192223 UP000009046 UP000002358 UP000008237 UP000005205 UP000015103 UP000053825 UP000078542 UP000037069 UP000069940 UP000249989 UP000092461 UP000008820 UP000215335 UP000242457 UP000183832 UP000030765 UP000005203 UP000235965 UP000092460 UP000078200 UP000092444 UP000092443 UP000002320 UP000069272 UP000000673 UP000095301 UP000001819 UP000075880 UP000075900 UP000091820 UP000075883 UP000000803 UP000192221 UP000008711 UP000002282 UP000075886 UP000076408 UP000075882 UP000075903 UP000283509 UP000007062 UP000075902 UP000008792 UP000075840 UP000268350 UP000076407 UP000075920 UP000075885
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9IVJ2
A0A194PEN4
A0A2W1BVE6
A0A2A4JGT3
A0A194QYE1
A0A212EZB6
+ More
A0A3S2LKJ5 A0A0L7LB88 A0A2H1WTE2 A0A1W4X609 A0A1W4X5V5 A0A1W4X5V0 E0VBV5 K7IXE7 E2C5M6 A0A1Y1KXZ8 A0A348G604 A0A158NN55 A0A0P4W4J7 T1I808 A0A146KL11 A0A0A9XS95 A0A0K8SCK7 A0A069DX38 A0A023EZT7 A0A0V0G4N0 A0A0C9RFV4 A0A0C9Q8K2 A0A0L7R316 A0A195C295 A0A1L8E265 A0A1L8E290 A0A0L0CEC5 A0A182H1M1 A0A1B0CY21 Q16RL5 T1DG95 A0A1L8DTH7 A0A0K2UJ39 U5EWK1 A0A232FMA1 A0A2A3E6J8 A0A336K9H3 A0A1J1J458 A0A084WGL0 A0A087ZUP7 A0A1B6DL67 A0A2J7Q7T3 A0A1B0AVV4 A0A1B6EG93 A0A1A9VFE3 A0A1B0G9Y4 A0A1A9XR98 B0WYN8 A0A2M4BE23 A0A182FBI4 A0A2M4BE95 W5J3Y4 A0A1I8MPC7 A0A0R3P7Z3 A0A2M4BDC8 A0A0R3P7F9 A0A182JLE1 A0A1I8MPC6 A0A182RAP7 A0A1A9WD61 A0A182MBV6 M9PCH6 A0A2M4CIB2 A0A1W4U9C0 Q9VUH1 A0A2M3ZFZ0 A0A0Q5U713 A0A2M4A0T9 A0A2M4A0X2 A0A2M3YZQ0 A0A0R1E3Q4 A0A182Q376 A0A2M3YZQ4 A0A182Y621 A0A2M4A0V0 A0A0R3P300 A0A182LP15 A0A182URD4 A0A3R7LWT5 A0A1S4H707 Q7PSF9 A0A182UKA9 B4LF51 A0A182HN75 A0A1W4UKP7 A0A3B0J3T2 A0A0Q5UB85 Q29R16 A0A182X7R6 A0A182WEQ8 A0A182PUL1 A0A0R1DY35 A0A0A1X6T7 A0A0J9RVJ9
A0A3S2LKJ5 A0A0L7LB88 A0A2H1WTE2 A0A1W4X609 A0A1W4X5V5 A0A1W4X5V0 E0VBV5 K7IXE7 E2C5M6 A0A1Y1KXZ8 A0A348G604 A0A158NN55 A0A0P4W4J7 T1I808 A0A146KL11 A0A0A9XS95 A0A0K8SCK7 A0A069DX38 A0A023EZT7 A0A0V0G4N0 A0A0C9RFV4 A0A0C9Q8K2 A0A0L7R316 A0A195C295 A0A1L8E265 A0A1L8E290 A0A0L0CEC5 A0A182H1M1 A0A1B0CY21 Q16RL5 T1DG95 A0A1L8DTH7 A0A0K2UJ39 U5EWK1 A0A232FMA1 A0A2A3E6J8 A0A336K9H3 A0A1J1J458 A0A084WGL0 A0A087ZUP7 A0A1B6DL67 A0A2J7Q7T3 A0A1B0AVV4 A0A1B6EG93 A0A1A9VFE3 A0A1B0G9Y4 A0A1A9XR98 B0WYN8 A0A2M4BE23 A0A182FBI4 A0A2M4BE95 W5J3Y4 A0A1I8MPC7 A0A0R3P7Z3 A0A2M4BDC8 A0A0R3P7F9 A0A182JLE1 A0A1I8MPC6 A0A182RAP7 A0A1A9WD61 A0A182MBV6 M9PCH6 A0A2M4CIB2 A0A1W4U9C0 Q9VUH1 A0A2M3ZFZ0 A0A0Q5U713 A0A2M4A0T9 A0A2M4A0X2 A0A2M3YZQ0 A0A0R1E3Q4 A0A182Q376 A0A2M3YZQ4 A0A182Y621 A0A2M4A0V0 A0A0R3P300 A0A182LP15 A0A182URD4 A0A3R7LWT5 A0A1S4H707 Q7PSF9 A0A182UKA9 B4LF51 A0A182HN75 A0A1W4UKP7 A0A3B0J3T2 A0A0Q5UB85 Q29R16 A0A182X7R6 A0A182WEQ8 A0A182PUL1 A0A0R1DY35 A0A0A1X6T7 A0A0J9RVJ9
PDB
1Z68
E-value=3.06519e-89,
Score=840
Ontologies
KEGG
GO
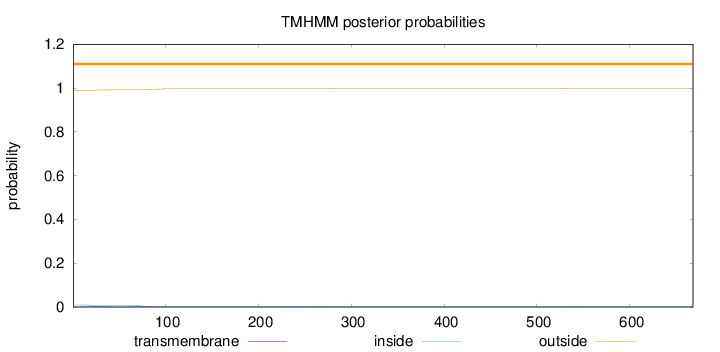
Topology
Length:
668
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16803
Exp number, first 60 AAs:
0.03863
Total prob of N-in:
0.00961
outside
1 - 668
Population Genetic Test Statistics
Pi
314.663273
Theta
179.298223
Tajima's D
1.901593
CLR
0.392825
CSRT
0.868256587170641
Interpretation
Uncertain
