Gene
KWMTBOMO07700
Pre Gene Modal
BGIBMGA001273
Annotation
PREDICTED:_trimethyllysine_dioxygenase?_mitochondrial_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.496
Sequence
CDS
ATGGAATTCAGTGTCGGATTCAATGAAAACAGTGCGAAAAAAATAGAAAACGTCATCTTTGTTGAGAGAACGGTACGTGTGAACTTCGAGAATGGCCCTTCGATTCCCTTTGAAGACTGTTGGCTCAGAGACCATTGCAGATGTTCACAATGCTATCACGCGAATACGTTCCAGAGAGCTAAACACATTTTAGAACTGCCCGATTCGAAGATACTTACATTACAATTCGATAAGAACAGTCTAACAATTGAATGGGACGATAAGCACACTACAGAATTCAAAGCGGACTTTCTGTCGCAATTCGATTATAAAACTTGGAAAAACAATCGCAGATTGAAGCCTAGATTGTGGCGCGGCTGTAACGTTGCTGATAGAATAGCTAAAGTACACGTTGATGAGTTTCTTGACTCTGACGACAGTTCAAAGGAAGTATTTCAATCGCTTCTTGACTATGGCGTTGCCTTTATAACTGGTGTGCAACCAAGCGCGGAAGCTACAGAAACAGTGTGCAAAGCTTTAGGAGGAATTCAACACACTATTTTTGGTGCTACGTGGGAATTCACCACAGTTGCTGATCACGCCGACACGGCCTATACCAATCTACCTTTGGCTGCGCACAATGATAATATTTATTGGACTGAAGCGGCTGGCCTCCAAATCCTTCACTGCATAGAACATACAAACGGTACGGGCGGTGAAACTATTCTTGTTGACGGCTTCTACGGAGCGACTTGTCTGAAGGAAGATCACCCGGAGGATTATGAATTCCTAACAACATACGAGATTGAGGGCGAATATATAGAACGTCGTCATCATTTCACGCATAGCGCACCTGTTATCCAAATCGACAAGAATACTGAAGACATTAAACAAATAAGATTCAATGTATACGATAGATCAGCTATGGCATTCAGGAGTGGCAGGGACTGTCGGCTCTACTATCGTTCCTTGAAAAATCTAGCTCGGTATTACGAAAATAAGGAAAACCAATGGATATTCAAATTAGTTCCAGGTCTTGTGATGGTAATAGACAACTTCAGATTGCTTCACGGTAGAAACGGTTTCACTGGAAGGCGGGTGCTGTGCGGTGCGTATGTGTCGCGCTCTGACTGGCTGGACAAAGCTCGGTCGTTAAATCTCATTCAATAA
Protein
MEFSVGFNENSAKKIENVIFVERTVRVNFENGPSIPFEDCWLRDHCRCSQCYHANTFQRAKHILELPDSKILTLQFDKNSLTIEWDDKHTTEFKADFLSQFDYKTWKNNRRLKPRLWRGCNVADRIAKVHVDEFLDSDDSSKEVFQSLLDYGVAFITGVQPSAEATETVCKALGGIQHTIFGATWEFTTVADHADTAYTNLPLAAHNDNIYWTEAAGLQILHCIEHTNGTGGETILVDGFYGATCLKEDHPEDYEFLTTYEIEGEYIERRHHFTHSAPVIQIDKNTEDIKQIRFNVYDRSAMAFRSGRDCRLYYRSLKNLARYYENKENQWIFKLVPGLVMVIDNFRLLHGRNGFTGRRVLCGAYVSRSDWLDKARSLNLIQ
Summary
Uniprot
A0A2H1W362
A0A2A4IYX8
A0A2W1BNY7
A0A212EQS3
A0A194R3Y5
E0W3Z1
+ More
A0A2J7PIE2 A0A1W4VAC9 A0A0P8XDL4 Q4V6C2 Q9VDM7 A0A067RHL2 A0A3B0JK47 A0A2M4AUV5 B4PP89 B4NFY2 B3NYY8 Q7Q783 B4G3E3 B4QS38 A0A2C9GQR7 A0A1A9V187 A0A182H2U4 A0A1A9X281 Q298C1 W5JCZ6 A0A182P3E3 A0A1B0GAW2 B4JXU3 A0A1A9WXT9 A0A084VDL8 B4IIC8 A0A034WQ21 A0A182FHS4 A0A1B0CUK0 A0A1A9YHK4 A0A0K8UBI8 A0A1B0C5L4 W8BX80 B4K7S6 A0A1B0A855 B4LVR9 A0A182UY60 A0A182Y0T1 A0A182LJR7 A0A182SDK9 Q16V01 A0A182IYB6 A0A182TCU2 A0A1I8P1R2 A0A182XCT9 A0A336LY45 A0A1W4X4U9 A0A0L0CG61 T1P8F3 A0A182K094 A0A1I8NH67 A0A182NMB1 A0A182RC05 A0A182QT31 A0A1Q3G1R0 A0A1B6EFC3 A0A182WLW0 K1QTC9 Q4V6P2 A0A0M5J2K7 B5X2I0 A0A1Y1MIG2 A7SLB9 A0A3N0XVV5 A0A060WTM1 A0A1S3RJJ3 A0A1Y1MIC5 A0A3M6UGA4 E9GXY5 A0A1S3L087 A0A1S3L1E6 A0A1B6I818 A0A167R729 A0A3Q7VS44 A0A3P9N6H8 A0A2Y9GJX7 E7F0M9 W5N8E5 G1SVM2 A0A3B0KA97 M3YXG0 A0A2U3XZW7 A0A3P9N6H4 B0WYB5 U6CRA1 D2I4U4 A0A3Q7NEI3 A0A2Y9IEH9 A0A2U3X1X8 A0A1U7STW9 A0A087YMJ4
A0A2J7PIE2 A0A1W4VAC9 A0A0P8XDL4 Q4V6C2 Q9VDM7 A0A067RHL2 A0A3B0JK47 A0A2M4AUV5 B4PP89 B4NFY2 B3NYY8 Q7Q783 B4G3E3 B4QS38 A0A2C9GQR7 A0A1A9V187 A0A182H2U4 A0A1A9X281 Q298C1 W5JCZ6 A0A182P3E3 A0A1B0GAW2 B4JXU3 A0A1A9WXT9 A0A084VDL8 B4IIC8 A0A034WQ21 A0A182FHS4 A0A1B0CUK0 A0A1A9YHK4 A0A0K8UBI8 A0A1B0C5L4 W8BX80 B4K7S6 A0A1B0A855 B4LVR9 A0A182UY60 A0A182Y0T1 A0A182LJR7 A0A182SDK9 Q16V01 A0A182IYB6 A0A182TCU2 A0A1I8P1R2 A0A182XCT9 A0A336LY45 A0A1W4X4U9 A0A0L0CG61 T1P8F3 A0A182K094 A0A1I8NH67 A0A182NMB1 A0A182RC05 A0A182QT31 A0A1Q3G1R0 A0A1B6EFC3 A0A182WLW0 K1QTC9 Q4V6P2 A0A0M5J2K7 B5X2I0 A0A1Y1MIG2 A7SLB9 A0A3N0XVV5 A0A060WTM1 A0A1S3RJJ3 A0A1Y1MIC5 A0A3M6UGA4 E9GXY5 A0A1S3L087 A0A1S3L1E6 A0A1B6I818 A0A167R729 A0A3Q7VS44 A0A3P9N6H8 A0A2Y9GJX7 E7F0M9 W5N8E5 G1SVM2 A0A3B0KA97 M3YXG0 A0A2U3XZW7 A0A3P9N6H4 B0WYB5 U6CRA1 D2I4U4 A0A3Q7NEI3 A0A2Y9IEH9 A0A2U3X1X8 A0A1U7STW9 A0A087YMJ4
Pubmed
28756777
22118469
26354079
20566863
17994087
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 17550304 12364791 26483478 15632085 20920257 23761445 24438588 25348373 24495485 25244985 20966253 17510324 26108605 25315136 22992520 20433749 28004739 17615350 24755649 30382153 21292972 26659563 23594743 21993624 20010809
12537568 12537572 12537573 12537574 16110336 17569856 17569867 24845553 17550304 12364791 26483478 15632085 20920257 23761445 24438588 25348373 24495485 25244985 20966253 17510324 26108605 25315136 22992520 20433749 28004739 17615350 24755649 30382153 21292972 26659563 23594743 21993624 20010809
EMBL
ODYU01006015
SOQ47488.1
NWSH01004513
PCG65025.1
KZ149946
PZC76762.1
+ More
AGBW02013240 OWR43840.1 KQ460947 KPJ10551.1 DS235885 EEB20347.1 NEVH01025127 PNF16084.1 CH902623 KPU72769.1 BT022384 AAY54800.1 AE014297 AAF55763.1 KK852463 KDR23351.1 OUUW01000007 SPP82595.1 GGFK01011248 MBW44569.1 CM000160 EDW96128.1 CH964251 EDW83199.1 CH954181 EDV48391.1 AAAB01008960 EAA11960.4 CH479179 EDW24325.1 CM000364 EDX12234.1 JXUM01105999 KQ565014 KXJ71440.1 CM000070 EAL28034.2 ADMH02001518 ETN62222.1 CCAG010014940 CH916377 EDV90505.1 ATLV01011630 KE524705 KFB36062.1 CH480842 EDW49672.1 GAKP01002505 JAC56447.1 AJWK01029292 GDHF01031068 GDHF01028305 JAI21246.1 JAI24009.1 JXJN01026063 GAMC01008659 JAB97896.1 CH933806 EDW16447.1 CH940650 EDW67524.1 CH477607 EAT38365.1 UFQT01000177 SSX21187.1 JRES01000438 KNC31240.1 KA645001 AFP59630.1 AXCN02000936 GFDL01001313 JAV33732.1 GEDC01000671 JAS36627.1 JH815999 EKC24856.1 BT022264 AAY54680.1 CP012526 ALC47511.1 BT045249 ACI33511.1 GEZM01030645 JAV85451.1 DS469697 EDO35490.1 RJVU01059915 ROJ62483.1 FR904732 CDQ70703.1 GEZM01030637 JAV85464.1 RCHS01001596 RMX52683.1 GL732574 EFX75501.1 GECU01024659 JAS83047.1 KV417269 KZP00626.1 CR792441 AHAT01007388 SPP82596.1 AEYP01096475 AEYP01096476 AEYP01096477 AEYP01096478 DS232186 EDS36992.1 HAAF01001359 CCP73185.1 ACTA01108118 ACTA01116118 ACTA01124118 ACTA01132118 ACTA01140118 ACTA01148117 ACTA01156116 ACTA01164115 ACTA01172113 ACTA01180111 GL194550 EFB20721.1 AYCK01001996
AGBW02013240 OWR43840.1 KQ460947 KPJ10551.1 DS235885 EEB20347.1 NEVH01025127 PNF16084.1 CH902623 KPU72769.1 BT022384 AAY54800.1 AE014297 AAF55763.1 KK852463 KDR23351.1 OUUW01000007 SPP82595.1 GGFK01011248 MBW44569.1 CM000160 EDW96128.1 CH964251 EDW83199.1 CH954181 EDV48391.1 AAAB01008960 EAA11960.4 CH479179 EDW24325.1 CM000364 EDX12234.1 JXUM01105999 KQ565014 KXJ71440.1 CM000070 EAL28034.2 ADMH02001518 ETN62222.1 CCAG010014940 CH916377 EDV90505.1 ATLV01011630 KE524705 KFB36062.1 CH480842 EDW49672.1 GAKP01002505 JAC56447.1 AJWK01029292 GDHF01031068 GDHF01028305 JAI21246.1 JAI24009.1 JXJN01026063 GAMC01008659 JAB97896.1 CH933806 EDW16447.1 CH940650 EDW67524.1 CH477607 EAT38365.1 UFQT01000177 SSX21187.1 JRES01000438 KNC31240.1 KA645001 AFP59630.1 AXCN02000936 GFDL01001313 JAV33732.1 GEDC01000671 JAS36627.1 JH815999 EKC24856.1 BT022264 AAY54680.1 CP012526 ALC47511.1 BT045249 ACI33511.1 GEZM01030645 JAV85451.1 DS469697 EDO35490.1 RJVU01059915 ROJ62483.1 FR904732 CDQ70703.1 GEZM01030637 JAV85464.1 RCHS01001596 RMX52683.1 GL732574 EFX75501.1 GECU01024659 JAS83047.1 KV417269 KZP00626.1 CR792441 AHAT01007388 SPP82596.1 AEYP01096475 AEYP01096476 AEYP01096477 AEYP01096478 DS232186 EDS36992.1 HAAF01001359 CCP73185.1 ACTA01108118 ACTA01116118 ACTA01124118 ACTA01132118 ACTA01140118 ACTA01148117 ACTA01156116 ACTA01164115 ACTA01172113 ACTA01180111 GL194550 EFB20721.1 AYCK01001996
Proteomes
UP000218220
UP000007151
UP000053240
UP000009046
UP000235965
UP000192221
+ More
UP000007801 UP000000803 UP000027135 UP000268350 UP000002282 UP000007798 UP000008711 UP000007062 UP000008744 UP000000304 UP000078200 UP000069940 UP000249989 UP000091820 UP000001819 UP000000673 UP000075885 UP000092444 UP000001070 UP000030765 UP000001292 UP000069272 UP000092461 UP000092443 UP000092460 UP000009192 UP000092445 UP000008792 UP000075903 UP000076408 UP000075882 UP000075901 UP000008820 UP000075880 UP000075902 UP000095300 UP000076407 UP000192223 UP000037069 UP000075881 UP000095301 UP000075884 UP000075900 UP000075886 UP000075920 UP000005408 UP000092553 UP000001593 UP000193380 UP000087266 UP000275408 UP000000305 UP000076738 UP000286642 UP000242638 UP000248481 UP000000437 UP000018468 UP000001811 UP000000715 UP000245341 UP000002320 UP000008912 UP000286641 UP000248482 UP000245340 UP000189704 UP000028760
UP000007801 UP000000803 UP000027135 UP000268350 UP000002282 UP000007798 UP000008711 UP000007062 UP000008744 UP000000304 UP000078200 UP000069940 UP000249989 UP000091820 UP000001819 UP000000673 UP000075885 UP000092444 UP000001070 UP000030765 UP000001292 UP000069272 UP000092461 UP000092443 UP000092460 UP000009192 UP000092445 UP000008792 UP000075903 UP000076408 UP000075882 UP000075901 UP000008820 UP000075880 UP000075902 UP000095300 UP000076407 UP000192223 UP000037069 UP000075881 UP000095301 UP000075884 UP000075900 UP000075886 UP000075920 UP000005408 UP000092553 UP000001593 UP000193380 UP000087266 UP000275408 UP000000305 UP000076738 UP000286642 UP000242638 UP000248481 UP000000437 UP000018468 UP000001811 UP000000715 UP000245341 UP000002320 UP000008912 UP000286641 UP000248482 UP000245340 UP000189704 UP000028760
Interpro
SUPFAM
SSF110035
SSF110035
Gene 3D
ProteinModelPortal
A0A2H1W362
A0A2A4IYX8
A0A2W1BNY7
A0A212EQS3
A0A194R3Y5
E0W3Z1
+ More
A0A2J7PIE2 A0A1W4VAC9 A0A0P8XDL4 Q4V6C2 Q9VDM7 A0A067RHL2 A0A3B0JK47 A0A2M4AUV5 B4PP89 B4NFY2 B3NYY8 Q7Q783 B4G3E3 B4QS38 A0A2C9GQR7 A0A1A9V187 A0A182H2U4 A0A1A9X281 Q298C1 W5JCZ6 A0A182P3E3 A0A1B0GAW2 B4JXU3 A0A1A9WXT9 A0A084VDL8 B4IIC8 A0A034WQ21 A0A182FHS4 A0A1B0CUK0 A0A1A9YHK4 A0A0K8UBI8 A0A1B0C5L4 W8BX80 B4K7S6 A0A1B0A855 B4LVR9 A0A182UY60 A0A182Y0T1 A0A182LJR7 A0A182SDK9 Q16V01 A0A182IYB6 A0A182TCU2 A0A1I8P1R2 A0A182XCT9 A0A336LY45 A0A1W4X4U9 A0A0L0CG61 T1P8F3 A0A182K094 A0A1I8NH67 A0A182NMB1 A0A182RC05 A0A182QT31 A0A1Q3G1R0 A0A1B6EFC3 A0A182WLW0 K1QTC9 Q4V6P2 A0A0M5J2K7 B5X2I0 A0A1Y1MIG2 A7SLB9 A0A3N0XVV5 A0A060WTM1 A0A1S3RJJ3 A0A1Y1MIC5 A0A3M6UGA4 E9GXY5 A0A1S3L087 A0A1S3L1E6 A0A1B6I818 A0A167R729 A0A3Q7VS44 A0A3P9N6H8 A0A2Y9GJX7 E7F0M9 W5N8E5 G1SVM2 A0A3B0KA97 M3YXG0 A0A2U3XZW7 A0A3P9N6H4 B0WYB5 U6CRA1 D2I4U4 A0A3Q7NEI3 A0A2Y9IEH9 A0A2U3X1X8 A0A1U7STW9 A0A087YMJ4
A0A2J7PIE2 A0A1W4VAC9 A0A0P8XDL4 Q4V6C2 Q9VDM7 A0A067RHL2 A0A3B0JK47 A0A2M4AUV5 B4PP89 B4NFY2 B3NYY8 Q7Q783 B4G3E3 B4QS38 A0A2C9GQR7 A0A1A9V187 A0A182H2U4 A0A1A9X281 Q298C1 W5JCZ6 A0A182P3E3 A0A1B0GAW2 B4JXU3 A0A1A9WXT9 A0A084VDL8 B4IIC8 A0A034WQ21 A0A182FHS4 A0A1B0CUK0 A0A1A9YHK4 A0A0K8UBI8 A0A1B0C5L4 W8BX80 B4K7S6 A0A1B0A855 B4LVR9 A0A182UY60 A0A182Y0T1 A0A182LJR7 A0A182SDK9 Q16V01 A0A182IYB6 A0A182TCU2 A0A1I8P1R2 A0A182XCT9 A0A336LY45 A0A1W4X4U9 A0A0L0CG61 T1P8F3 A0A182K094 A0A1I8NH67 A0A182NMB1 A0A182RC05 A0A182QT31 A0A1Q3G1R0 A0A1B6EFC3 A0A182WLW0 K1QTC9 Q4V6P2 A0A0M5J2K7 B5X2I0 A0A1Y1MIG2 A7SLB9 A0A3N0XVV5 A0A060WTM1 A0A1S3RJJ3 A0A1Y1MIC5 A0A3M6UGA4 E9GXY5 A0A1S3L087 A0A1S3L1E6 A0A1B6I818 A0A167R729 A0A3Q7VS44 A0A3P9N6H8 A0A2Y9GJX7 E7F0M9 W5N8E5 G1SVM2 A0A3B0KA97 M3YXG0 A0A2U3XZW7 A0A3P9N6H4 B0WYB5 U6CRA1 D2I4U4 A0A3Q7NEI3 A0A2Y9IEH9 A0A2U3X1X8 A0A1U7STW9 A0A087YMJ4
PDB
4CWD
E-value=4.21917e-31,
Score=336
Ontologies
GO
PANTHER
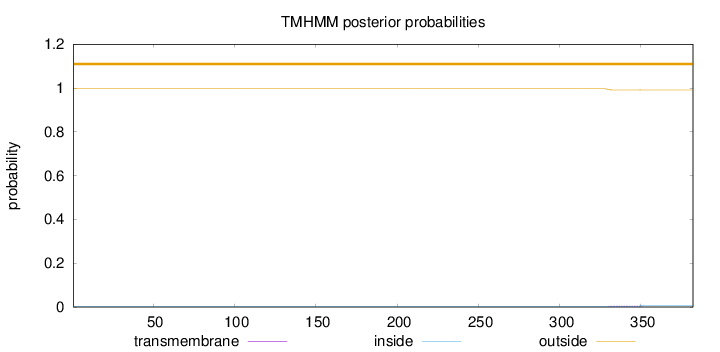
Topology
Length:
382
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11124
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00365
outside
1 - 382
Population Genetic Test Statistics
Pi
19.964456
Theta
169.18961
Tajima's D
-1.867988
CLR
1.494416
CSRT
0.0233988300584971
Interpretation
Uncertain
