Gene
KWMTBOMO07695
Pre Gene Modal
BGIBMGA001275
Annotation
PREDICTED:_carboxypeptidase_Q-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.93
Sequence
CDS
ATGTATGAAGATTATTCTAAGTTCATAGATACATTTGGAGCACGTTTGGCGGGAAGTAAAGCTCTAGAAAAAGCTATAGATCACATGGTAAATCTGACACGACAAAATGGCTTGAACGACGTGACAACGGAAGAAGTTGACATACCATACTGGCAACGTATTTACGAATCGGCAAACATGTTCGAACCATATTTAAAAAAAATTTCAATCCTTGGGTTAGGAACTAGCGTGAGCACTCCTGCTGAGGGTATCAAAGCAGAAGCCATTGTAATAAAAAGTTTTGACGAACTTGAAAAAACTGATGAAAATAAAATAAAAGGGAAAATTGTCGTCTTTGATCCACCTTTTGTGAGTTATGGTGAAACTGTTGTTTATAGAAGTCTAGGAGCTTCAAAGGCTTCCAGAAAAGGTGCTGTAGCTACTTTAGTCCGAAGCATTACACCATTCTCTATATATTCTCTTCACACCGGAGCCCAGGACTATGAAAATAATGTTACAAAAATACCTACTGCAGCTATAACTCACGAAGACGCAGACTTATTTAGAAGACTTCAAAATCTTGGCGAAAAAATAGTCCTTAAAATTCATATGTCTTATGCTATGTCCAGAAAGATATCAAGGAATACCATTATTGATTTAAAAGGCAAAGAAAGCCCCGAAAAACAAGTTATCGTGTCAGGACACATTGATAGCTGGGATGTAGGCCAGGGCGCTATGGATGACGGTGGGGGTATGATGATAAGTTGGTTCGTTCCTGTAATACTGAACATTTTAAAACTAAAACCAAGACGAACAATCCGAGCGATACTATGGACTGCAGAAGAGCCCGGTCTGATAGGTGCTGAAGCATATCTAACAAAACACATTAACGAAACTTCAAATATAAATTTTATCATGGAATCTGATGAAGGGACGTTCAAGCCTCTAGGGTTAGATGTCGCCGGCAGTACCAATGCTCGTTGCATTATAGCTGAAGTACTGAAACTGTTTACACCCATTGATAAAATGAAGGAAAGTGAATATCCGGGCTCGGATATGACGTTATTCGTGCAAAAGGGAATTCCCGGGGCATCATTATTGAACGAGAATGAGCGTTACTTTTGGTTTCATCACAGTGAAGGTGACACAATGAACGTACAAAATCAAGAGGACGTAGTTGCTTGTGCCGCGTTCTGGGCGGCTTTTTCGTATGTCATTGCGGATTTGTCACAAGATATACCACGACAATAA
Protein
MYEDYSKFIDTFGARLAGSKALEKAIDHMVNLTRQNGLNDVTTEEVDIPYWQRIYESANMFEPYLKKISILGLGTSVSTPAEGIKAEAIVIKSFDELEKTDENKIKGKIVVFDPPFVSYGETVVYRSLGASKASRKGAVATLVRSITPFSIYSLHTGAQDYENNVTKIPTAAITHEDADLFRRLQNLGEKIVLKIHMSYAMSRKISRNTIIDLKGKESPEKQVIVSGHIDSWDVGQGAMDDGGGMMISWFVPVILNILKLKPRRTIRAILWTAEEPGLIGAEAYLTKHINETSNINFIMESDEGTFKPLGLDVAGSTNARCIIAEVLKLFTPIDKMKESEYPGSDMTLFVQKGIPGASLLNENERYFWFHHSEGDTMNVQNQEDVVACAAFWAAFSYVIADLSQDIPRQ
Summary
Uniprot
H9IVJ5
A0A2W1BT05
A0A2A4JBH4
A0A2H1W0X3
A0A194PDQ8
A0A194QYM1
+ More
I6QPL8 A0A0L7KU87 A0A0L7L507 A0A212EKK8 A0A3S2LQH9 A0A194PD42 A0A2H1X2C5 A0A2A4IU93 A0A2W1BYC3 A0A194R3Z0 H9IVJ4 A0A2A4ITH1 A0A212F1M6 A0A1W4WA03 A0A1B6D5B2 A0A0P6GX37 A0A0P5ZCG3 A0A0N8CLN8 A0A0P6ELZ7 A0A0P6FUR6 A0A0P5TUH7 A0A0P6EJ76 A0A0P4YP03 A0A0P6GPG4 A0A0P5YA43 A0A0P5YDD7 A0A0P5YA68 A0A0P4ZHU5 A0A0N7ZPA7 A0A0P6FHL4 A0A0E9X5R7 A0A0P5PD60 A0A0P6ESB3 A0A0P6EPI2 A0A0P4YFZ9 J3JUI5 A0A0P6G5F7 A0A0P6I966 A0A0P5HJK5 A0A0P5HB97 A0A0P5VAJ5 A0A0P5H3Z1 A0A0P5J438 E9HGJ7 C3XQU6 A0A1D2N881 N6UW91 A0A3Q3GGS9 K7IXP8 A0A3B4F9T0 A0A3Q3B5Z6 A0A3P8ZHX7 A0A2U9AYY7 A0A3Q2VHV9 A0A0P6GE38 A0A3Q1KFW5 A0A3Q1CG28 A0A1A8L420 A0A1A8N8C4 A0A3Q1H2B3 A0A3P8PR29 A0A3P9DHC2 I3JIC2 A0A1A8BUE3 A0A1A8UMC0 A0A1A7ZT19 A0A1A8DSW8 V5I8M9 A0A3B4BI73 A0A3P8TST6 H2U0H2 A0A3P8PRF1 A0A3P8UXC3 A0A3P8TX70 H3CW82 A0A3P8UXC1 A0A3R7MAF5 A0A1A7WS62 A0A3B3BXV5 A0A3B5KCH1 A0A146NDV3 A0A3Q2DD90 A0A2I4CEE0 A0A2D0RHU4 B5X4A1 A0A1S5QN55 A0A2D0RGP5 A0A154P9F5 A0A146RWP5 A0A1I9WL52 A0A3B5A923 A0A3B5MKA4 A0A3Q2Q6Q3 A0A3Q2NQB3
I6QPL8 A0A0L7KU87 A0A0L7L507 A0A212EKK8 A0A3S2LQH9 A0A194PD42 A0A2H1X2C5 A0A2A4IU93 A0A2W1BYC3 A0A194R3Z0 H9IVJ4 A0A2A4ITH1 A0A212F1M6 A0A1W4WA03 A0A1B6D5B2 A0A0P6GX37 A0A0P5ZCG3 A0A0N8CLN8 A0A0P6ELZ7 A0A0P6FUR6 A0A0P5TUH7 A0A0P6EJ76 A0A0P4YP03 A0A0P6GPG4 A0A0P5YA43 A0A0P5YDD7 A0A0P5YA68 A0A0P4ZHU5 A0A0N7ZPA7 A0A0P6FHL4 A0A0E9X5R7 A0A0P5PD60 A0A0P6ESB3 A0A0P6EPI2 A0A0P4YFZ9 J3JUI5 A0A0P6G5F7 A0A0P6I966 A0A0P5HJK5 A0A0P5HB97 A0A0P5VAJ5 A0A0P5H3Z1 A0A0P5J438 E9HGJ7 C3XQU6 A0A1D2N881 N6UW91 A0A3Q3GGS9 K7IXP8 A0A3B4F9T0 A0A3Q3B5Z6 A0A3P8ZHX7 A0A2U9AYY7 A0A3Q2VHV9 A0A0P6GE38 A0A3Q1KFW5 A0A3Q1CG28 A0A1A8L420 A0A1A8N8C4 A0A3Q1H2B3 A0A3P8PR29 A0A3P9DHC2 I3JIC2 A0A1A8BUE3 A0A1A8UMC0 A0A1A7ZT19 A0A1A8DSW8 V5I8M9 A0A3B4BI73 A0A3P8TST6 H2U0H2 A0A3P8PRF1 A0A3P8UXC3 A0A3P8TX70 H3CW82 A0A3P8UXC1 A0A3R7MAF5 A0A1A7WS62 A0A3B3BXV5 A0A3B5KCH1 A0A146NDV3 A0A3Q2DD90 A0A2I4CEE0 A0A2D0RHU4 B5X4A1 A0A1S5QN55 A0A2D0RGP5 A0A154P9F5 A0A146RWP5 A0A1I9WL52 A0A3B5A923 A0A3B5MKA4 A0A3Q2Q6Q3 A0A3Q2NQB3
Pubmed
EMBL
BABH01001228
KZ149946
PZC76765.1
NWSH01002099
PCG69199.1
ODYU01005659
+ More
SOQ46741.1 KQ459606 KPI91153.1 KQ460947 KPJ10557.1 JQ653044 AFM38216.1 JTDY01005618 KOB66823.1 JTDY01002866 KOB70573.1 AGBW02014249 OWR42016.1 RSAL01000457 RVE41646.1 KPI91152.1 ODYU01012931 SOQ59473.1 NWSH01007446 PCG62968.1 PZC76763.1 KPJ10556.1 BABH01001213 PCG62969.1 AGBW02010848 OWR47639.1 GEDC01016472 GEDC01000320 JAS20826.1 JAS36978.1 GDIQ01040118 JAN54619.1 GDIP01058450 GDIQ01042768 JAM45265.1 JAN51969.1 GDIP01119543 JAL84171.1 GDIP01122640 GDIQ01061715 JAL81074.1 JAN33022.1 GDIQ01042767 JAN51970.1 GDIQ01141482 GDIP01121438 GDIP01058451 JAL10244.1 JAL82276.1 GDIP01083545 GDIQ01061714 JAM20170.1 JAN33023.1 GDIP01225880 JAI97521.1 GDIP01201994 GDIQ01154770 GDIP01058452 GDIQ01051316 GDIQ01042769 LRGB01000512 JAJ21408.1 JAN51968.1 KZS18795.1 GDIP01060702 JAM43013.1 GDIP01060700 JAM43015.1 GDIP01060701 JAM43014.1 GDIP01225879 JAI97522.1 GDIP01225881 JAI97520.1 GDIQ01059661 JAN35076.1 GBXM01010500 JAH98077.1 GDIQ01151925 JAK99800.1 GDIQ01059662 JAN35075.1 GDIQ01059663 JAN35074.1 GDIP01228085 JAI95316.1 BT126899 AEE61861.1 GDIQ01049919 JAN44818.1 GDIQ01009486 JAN85251.1 GDIQ01232432 JAK19293.1 GDIQ01232434 JAK19291.1 GDIP01102645 JAM01070.1 GDIQ01232433 JAK19292.1 GDIQ01205514 JAK46211.1 GL732642 EFX69094.1 GG666456 EEN69541.1 LJIJ01000168 ODN01176.1 APGK01006246 APGK01006577 KB736835 KB736662 KB631984 ENN83126.1 ENN83167.1 ERL87685.1 CP026243 AWO96874.1 GDIQ01034566 JAN60171.1 HAEF01002000 SBR39382.1 HAEH01000670 HAEI01013854 SBR65343.1 AERX01027706 AERX01027707 AERX01027708 HADZ01007346 SBP71287.1 HAEJ01008279 SBS48736.1 HADY01006971 SBP45456.1 HAEA01007439 SBQ35919.1 GALX01004405 JAB64061.1 QCYY01001532 ROT77368.1 HADW01007091 HADX01011225 SBP08491.1 GCES01156710 JAQ29612.1 BT045870 ACI34132.1 KR068535 AMO02552.1 KQ434850 KZC08566.1 GCES01113519 JAQ72803.1 KU932236 APA33872.1
SOQ46741.1 KQ459606 KPI91153.1 KQ460947 KPJ10557.1 JQ653044 AFM38216.1 JTDY01005618 KOB66823.1 JTDY01002866 KOB70573.1 AGBW02014249 OWR42016.1 RSAL01000457 RVE41646.1 KPI91152.1 ODYU01012931 SOQ59473.1 NWSH01007446 PCG62968.1 PZC76763.1 KPJ10556.1 BABH01001213 PCG62969.1 AGBW02010848 OWR47639.1 GEDC01016472 GEDC01000320 JAS20826.1 JAS36978.1 GDIQ01040118 JAN54619.1 GDIP01058450 GDIQ01042768 JAM45265.1 JAN51969.1 GDIP01119543 JAL84171.1 GDIP01122640 GDIQ01061715 JAL81074.1 JAN33022.1 GDIQ01042767 JAN51970.1 GDIQ01141482 GDIP01121438 GDIP01058451 JAL10244.1 JAL82276.1 GDIP01083545 GDIQ01061714 JAM20170.1 JAN33023.1 GDIP01225880 JAI97521.1 GDIP01201994 GDIQ01154770 GDIP01058452 GDIQ01051316 GDIQ01042769 LRGB01000512 JAJ21408.1 JAN51968.1 KZS18795.1 GDIP01060702 JAM43013.1 GDIP01060700 JAM43015.1 GDIP01060701 JAM43014.1 GDIP01225879 JAI97522.1 GDIP01225881 JAI97520.1 GDIQ01059661 JAN35076.1 GBXM01010500 JAH98077.1 GDIQ01151925 JAK99800.1 GDIQ01059662 JAN35075.1 GDIQ01059663 JAN35074.1 GDIP01228085 JAI95316.1 BT126899 AEE61861.1 GDIQ01049919 JAN44818.1 GDIQ01009486 JAN85251.1 GDIQ01232432 JAK19293.1 GDIQ01232434 JAK19291.1 GDIP01102645 JAM01070.1 GDIQ01232433 JAK19292.1 GDIQ01205514 JAK46211.1 GL732642 EFX69094.1 GG666456 EEN69541.1 LJIJ01000168 ODN01176.1 APGK01006246 APGK01006577 KB736835 KB736662 KB631984 ENN83126.1 ENN83167.1 ERL87685.1 CP026243 AWO96874.1 GDIQ01034566 JAN60171.1 HAEF01002000 SBR39382.1 HAEH01000670 HAEI01013854 SBR65343.1 AERX01027706 AERX01027707 AERX01027708 HADZ01007346 SBP71287.1 HAEJ01008279 SBS48736.1 HADY01006971 SBP45456.1 HAEA01007439 SBQ35919.1 GALX01004405 JAB64061.1 QCYY01001532 ROT77368.1 HADW01007091 HADX01011225 SBP08491.1 GCES01156710 JAQ29612.1 BT045870 ACI34132.1 KR068535 AMO02552.1 KQ434850 KZC08566.1 GCES01113519 JAQ72803.1 KU932236 APA33872.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000283053 UP000192223 UP000076858 UP000000305 UP000001554 UP000094527 UP000019118 UP000030742 UP000261660 UP000002358 UP000261460 UP000264800 UP000265140 UP000246464 UP000264840 UP000265040 UP000257160 UP000257200 UP000265100 UP000265160 UP000005207 UP000261520 UP000265080 UP000005226 UP000265120 UP000007303 UP000283509 UP000261560 UP000265020 UP000192220 UP000221080 UP000087266 UP000076502 UP000261400 UP000261380 UP000265000
UP000283053 UP000192223 UP000076858 UP000000305 UP000001554 UP000094527 UP000019118 UP000030742 UP000261660 UP000002358 UP000261460 UP000264800 UP000265140 UP000246464 UP000264840 UP000265040 UP000257160 UP000257200 UP000265100 UP000265160 UP000005207 UP000261520 UP000265080 UP000005226 UP000265120 UP000007303 UP000283509 UP000261560 UP000265020 UP000192220 UP000221080 UP000087266 UP000076502 UP000261400 UP000261380 UP000265000
Interpro
ProteinModelPortal
H9IVJ5
A0A2W1BT05
A0A2A4JBH4
A0A2H1W0X3
A0A194PDQ8
A0A194QYM1
+ More
I6QPL8 A0A0L7KU87 A0A0L7L507 A0A212EKK8 A0A3S2LQH9 A0A194PD42 A0A2H1X2C5 A0A2A4IU93 A0A2W1BYC3 A0A194R3Z0 H9IVJ4 A0A2A4ITH1 A0A212F1M6 A0A1W4WA03 A0A1B6D5B2 A0A0P6GX37 A0A0P5ZCG3 A0A0N8CLN8 A0A0P6ELZ7 A0A0P6FUR6 A0A0P5TUH7 A0A0P6EJ76 A0A0P4YP03 A0A0P6GPG4 A0A0P5YA43 A0A0P5YDD7 A0A0P5YA68 A0A0P4ZHU5 A0A0N7ZPA7 A0A0P6FHL4 A0A0E9X5R7 A0A0P5PD60 A0A0P6ESB3 A0A0P6EPI2 A0A0P4YFZ9 J3JUI5 A0A0P6G5F7 A0A0P6I966 A0A0P5HJK5 A0A0P5HB97 A0A0P5VAJ5 A0A0P5H3Z1 A0A0P5J438 E9HGJ7 C3XQU6 A0A1D2N881 N6UW91 A0A3Q3GGS9 K7IXP8 A0A3B4F9T0 A0A3Q3B5Z6 A0A3P8ZHX7 A0A2U9AYY7 A0A3Q2VHV9 A0A0P6GE38 A0A3Q1KFW5 A0A3Q1CG28 A0A1A8L420 A0A1A8N8C4 A0A3Q1H2B3 A0A3P8PR29 A0A3P9DHC2 I3JIC2 A0A1A8BUE3 A0A1A8UMC0 A0A1A7ZT19 A0A1A8DSW8 V5I8M9 A0A3B4BI73 A0A3P8TST6 H2U0H2 A0A3P8PRF1 A0A3P8UXC3 A0A3P8TX70 H3CW82 A0A3P8UXC1 A0A3R7MAF5 A0A1A7WS62 A0A3B3BXV5 A0A3B5KCH1 A0A146NDV3 A0A3Q2DD90 A0A2I4CEE0 A0A2D0RHU4 B5X4A1 A0A1S5QN55 A0A2D0RGP5 A0A154P9F5 A0A146RWP5 A0A1I9WL52 A0A3B5A923 A0A3B5MKA4 A0A3Q2Q6Q3 A0A3Q2NQB3
I6QPL8 A0A0L7KU87 A0A0L7L507 A0A212EKK8 A0A3S2LQH9 A0A194PD42 A0A2H1X2C5 A0A2A4IU93 A0A2W1BYC3 A0A194R3Z0 H9IVJ4 A0A2A4ITH1 A0A212F1M6 A0A1W4WA03 A0A1B6D5B2 A0A0P6GX37 A0A0P5ZCG3 A0A0N8CLN8 A0A0P6ELZ7 A0A0P6FUR6 A0A0P5TUH7 A0A0P6EJ76 A0A0P4YP03 A0A0P6GPG4 A0A0P5YA43 A0A0P5YDD7 A0A0P5YA68 A0A0P4ZHU5 A0A0N7ZPA7 A0A0P6FHL4 A0A0E9X5R7 A0A0P5PD60 A0A0P6ESB3 A0A0P6EPI2 A0A0P4YFZ9 J3JUI5 A0A0P6G5F7 A0A0P6I966 A0A0P5HJK5 A0A0P5HB97 A0A0P5VAJ5 A0A0P5H3Z1 A0A0P5J438 E9HGJ7 C3XQU6 A0A1D2N881 N6UW91 A0A3Q3GGS9 K7IXP8 A0A3B4F9T0 A0A3Q3B5Z6 A0A3P8ZHX7 A0A2U9AYY7 A0A3Q2VHV9 A0A0P6GE38 A0A3Q1KFW5 A0A3Q1CG28 A0A1A8L420 A0A1A8N8C4 A0A3Q1H2B3 A0A3P8PR29 A0A3P9DHC2 I3JIC2 A0A1A8BUE3 A0A1A8UMC0 A0A1A7ZT19 A0A1A8DSW8 V5I8M9 A0A3B4BI73 A0A3P8TST6 H2U0H2 A0A3P8PRF1 A0A3P8UXC3 A0A3P8TX70 H3CW82 A0A3P8UXC1 A0A3R7MAF5 A0A1A7WS62 A0A3B3BXV5 A0A3B5KCH1 A0A146NDV3 A0A3Q2DD90 A0A2I4CEE0 A0A2D0RHU4 B5X4A1 A0A1S5QN55 A0A2D0RGP5 A0A154P9F5 A0A146RWP5 A0A1I9WL52 A0A3B5A923 A0A3B5MKA4 A0A3Q2Q6Q3 A0A3Q2NQB3
PDB
3IIB
E-value=3.08826e-45,
Score=458
Ontologies
GO
PANTHER
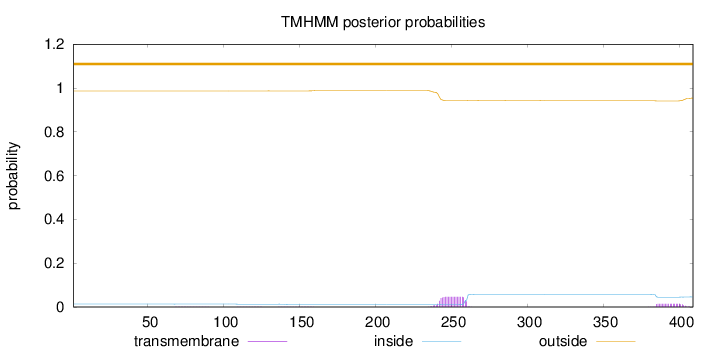
Topology
Length:
409
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.16688
Exp number, first 60 AAs:
1e-05
Total prob of N-in:
0.01346
outside
1 - 409
Population Genetic Test Statistics
Pi
197.314
Theta
183.768388
Tajima's D
0.227185
CLR
3.001605
CSRT
0.434428278586071
Interpretation
Uncertain
