Gene
KWMTBOMO07694
Pre Gene Modal
BGIBMGA001276
Annotation
PREDICTED:_cytochrome_P450_CYP12A2_isoform_X2_[Bombyx_mori]
Full name
Cytochrome P450 CYP12A2
Alternative Name
CYPXIIA2
Location in the cell
Mitochondrial Reliability : 1.394 Nuclear Reliability : 1.797
Sequence
CDS
ATGATAATAGCCCTGCATTACAGTAAACTGAGACCGGTGTTGAATTTCTCGTGTCTGCAGCAATGTGTGAGAACAGTGACAGTTAGCGCGGCAACAGAAAAACTACAACAAACAGAATTGAAATCGTTTCGAGAGATTCCGGGCCCCTCGTCGTTGCCGATCATGGGCCCGTTTCTACACTTCATGCCCGGAGGATCCCTTCACAACATTAACAGCACGGAACTGACTCACAAACTGTACGACATATACGGACCTATCGTTAGAATCGATTCAATGTTTTCTAAGGACGCTATTGTATTACTGTATGATGCAGAAAGCGCCGGAATCATTTTACGTAACGAGAACAACATGCCAATAAGGATCAGCTTTAAATCCTTATCATATTATAGACAGAAATATAAGAAATCGGAAAATGATCGCACAGACAGACCGACTGGATTAGTGTCAGACCACGGCGAGTTATGGAAGAGCTTCCGTTCGGCGGTGAATCCAGTCTTATTACAGCCGAAAACTATAAGACTATACAGCTCAGCACTAGAAGAGGTAGCCACCGACATGGTCGAGAGGCTTAGGTCTCTTAGAGATGAAAACAACAGGATACGTGGTCAATTCGATCAAGAGATGAATCTTTGGTCTCTCGAATCAATAGGGGTTGTGGCTTTAGGAAACCGGCTTAACTGCTTCGATTCAAATCTTCAGGACGACTCCCCCGTCAAGAGATTGATCGAATGTGTCCATCAGATGTTCGTGTTATCGAACGAATTGGACCTCAAGCCCAGTATTTGGACGTATGTTTCAACACCGCTATTCAGGAAAACTATGAAAATGTATGAAGAACAAGAAAATTTAACGAAATATTTCATCAAGAAAGCACTAGACGATATTAAAATGAACAAATCGAAATCGGATGATGAGAAACCGGTGCTCGAGAAATTATTGGACATCAATGAAGAATACGCGTATATTATGGCTTCAGACATGCTGGTGGCTGGTGTCGATACCACTTCAAACACGATGTCCGCAACTTTGTACCTCATGGCGATAAATCAGGACAAACAGCAGAAGTTGAGGGAAGAAGTGATGTCGAAGAACGGTAAAAGATCATACTTGAGGGCTTGCATCAAAGAGGCCATGAGAATACTACCAGTCGTGTCCGGGAACATGCGACGCACCACAAAGGAGTACAACATTCTTGGATATCATATACCGGAAAATGTCGACATTGCATTTGCTCATCAACATTTGTCGATGATGGAGAAATACTATCCAAGACCGACGGAGTTCATTCCTGAACGCTGGTTGACTAACAAGAGCGATCCTCTTTATTATGGAAACGCGCATCCATTCGCGAATTCCCCATTCGGCTTCGGTGTTCGCAGCTGTATAGGTCGTCGCATCGCCGAATTAGAAGTGGAAACATTTTTGTCGAAGATCGTCGAAAATTTTCAAGTCGAGTGGTCGGGGTCATCCCCTAGAGTCGAACAGACTTCCATCAATTACTTCAAGGGACCTTTTAACTTTATATTCAAAGATCTTTAA
Protein
MIIALHYSKLRPVLNFSCLQQCVRTVTVSAATEKLQQTELKSFREIPGPSSLPIMGPFLHFMPGGSLHNINSTELTHKLYDIYGPIVRIDSMFSKDAIVLLYDAESAGIILRNENNMPIRISFKSLSYYRQKYKKSENDRTDRPTGLVSDHGELWKSFRSAVNPVLLQPKTIRLYSSALEEVATDMVERLRSLRDENNRIRGQFDQEMNLWSLESIGVVALGNRLNCFDSNLQDDSPVKRLIECVHQMFVLSNELDLKPSIWTYVSTPLFRKTMKMYEEQENLTKYFIKKALDDIKMNKSKSDDEKPVLEKLLDINEEYAYIMASDMLVAGVDTTSNTMSATLYLMAINQDKQQKLREEVMSKNGKRSYLRACIKEAMRILPVVSGNMRRTTKEYNILGYHIPENVDIAFAHQHLSMMEKYYPRPTEFIPERWLTNKSDPLYYGNAHPFANSPFGFGVRSCIGRRIAELEVETFLSKIVENFQVEWSGSSPRVEQTSINYFKGPFNFIFKDL
Summary
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Keywords
Complete proteome
Heme
Iron
Metal-binding
Monooxygenase
Oxidoreductase
Feature
chain Cytochrome P450 CYP12A2
Uniprot
L0N5L2
L0N6M0
H9IVJ6
L0N731
D5L0N7
A0A068ETR9
+ More
D5L0N6 A0A2A4JB57 J7FIT7 A0A286QUH6 A0A248QEI8 A0A194PD37 A0A194PCV8 A0A0C5C533 A0A0C5C1H5 D2JLK5 A0A194QYD6 A0A1V0D9G9 A0A1V0D9G5 A0A212ERS8 H9IVJ7 A0A1E1WBR0 A0A1E1W6Z4 A0A1E1WJG1 A0A346II03 A0A212F2K7 A0A1V0D9I0 A0A194QYL5 A0A1E1WN58 A0A1E1WAC2 A0A194QZ45 A0A0N0PEW1 A0A068EWM5 A0A286MXN7 A0A194PNU6 J7FJF8 A0A212EU53 A0A248QEI9 A0A194QSH9 D5L0N5 L0N6I8 A0A3S2PF19 A0A0L7LAT5 A0A0C5C1L2 A0A182MHV7 A0A2H1WI68 A0A340TBF6 A0A1I8MP30 A0A336K3T6 A0A182PZ56 A0A023ETJ1 A0A182H3M9 O18635 A0A1S4GX15 K7J4S2 Q5TQM6 A0A182II69 A0A182UM32 A0A182XUR2 A0A182XUR0 A0A084VBX5 Q86QN3 A0A1S4GXQ5 Q17JN3 A0A1W6R7J5 Q86QN4 A0A182VHC9 A0A1L8E4A8 A0A182II71 A0A1L8E3Y1 A0A1L8E4J3 A0A1L8E4I2 A0A182S4Q4 A0A026X4P4 A0A182FZU6 A0A336MMT9 A0A2J7RNN8 A0A182PZ54 A0A1A9VZ41 B4KTY6 U5EHF6 A0A1Q3FJK3 A0A1Q3FR33 B0WTS5 A0A2M4AJC1 A0A1I8N7Q7 A0A182IJQ1 A0A2M4AI68 B0WTS3 A0A1S4JS48 A0A2J7PBF7 A0A068F5B4 A0A336MA77
D5L0N6 A0A2A4JB57 J7FIT7 A0A286QUH6 A0A248QEI8 A0A194PD37 A0A194PCV8 A0A0C5C533 A0A0C5C1H5 D2JLK5 A0A194QYD6 A0A1V0D9G9 A0A1V0D9G5 A0A212ERS8 H9IVJ7 A0A1E1WBR0 A0A1E1W6Z4 A0A1E1WJG1 A0A346II03 A0A212F2K7 A0A1V0D9I0 A0A194QYL5 A0A1E1WN58 A0A1E1WAC2 A0A194QZ45 A0A0N0PEW1 A0A068EWM5 A0A286MXN7 A0A194PNU6 J7FJF8 A0A212EU53 A0A248QEI9 A0A194QSH9 D5L0N5 L0N6I8 A0A3S2PF19 A0A0L7LAT5 A0A0C5C1L2 A0A182MHV7 A0A2H1WI68 A0A340TBF6 A0A1I8MP30 A0A336K3T6 A0A182PZ56 A0A023ETJ1 A0A182H3M9 O18635 A0A1S4GX15 K7J4S2 Q5TQM6 A0A182II69 A0A182UM32 A0A182XUR2 A0A182XUR0 A0A084VBX5 Q86QN3 A0A1S4GXQ5 Q17JN3 A0A1W6R7J5 Q86QN4 A0A182VHC9 A0A1L8E4A8 A0A182II71 A0A1L8E3Y1 A0A1L8E4J3 A0A1L8E4I2 A0A182S4Q4 A0A026X4P4 A0A182FZU6 A0A336MMT9 A0A2J7RNN8 A0A182PZ54 A0A1A9VZ41 B4KTY6 U5EHF6 A0A1Q3FJK3 A0A1Q3FR33 B0WTS5 A0A2M4AJC1 A0A1I8N7Q7 A0A182IJQ1 A0A2M4AI68 B0WTS3 A0A1S4JS48 A0A2J7PBF7 A0A068F5B4 A0A336MA77
EC Number
1.14.-.-
Pubmed
EMBL
AK343196
AK343202
BAM73885.1
BAM73888.1
AK343201
BAM73887.1
+ More
BABH01001230 BABH01001231 AK289309 BAM73836.1 GU731542 ADE05592.1 KM016710 KZ149946 AID54862.1 PZC76746.1 GU731541 ADE05591.1 NWSH01002099 PCG69201.1 JX310099 AFP20610.1 KX443481 ASO98056.1 KX443480 ASO98055.1 KQ459606 KPI91147.1 KPI91146.1 KP001115 AJN91160.1 KP001116 AJN91161.1 GQ915321 ACZ97415.1 KQ460947 KPJ10553.1 KY212080 ARA91644.1 KY212081 ARA91645.1 AGBW02012935 OWR44198.1 BABH01001232 GDQN01010473 GDQN01006630 JAT80581.1 JAT84424.1 GDQN01008311 JAT82743.1 GDQN01003905 JAT87149.1 MH138217 AXP17158.1 AGBW02010730 OWR47956.1 KY212082 ARA91646.1 KPJ10552.1 GDQN01002676 JAT88378.1 GDQN01007130 JAT83924.1 KPJ10554.1 KQ459668 KPJ20511.1 KM016709 AID54861.1 MF684355 ASX93989.1 KQ459599 KPI94419.1 JX310098 AFP20609.1 AGBW02012491 OWR44987.1 KX443479 ASO98054.1 KQ461154 KPJ08442.1 GU731540 ADE05590.1 AK289310 BAM73837.1 RSAL01000062 RVE49588.1 JTDY01001897 KOB72593.1 KP001114 AJN91159.1 AXCM01012394 ODYU01008822 SOQ52761.1 UFQS01000086 UFQT01000086 SSW99091.1 SSX19473.1 GAPW01000938 JAC12660.1 JXUM01024875 JXUM01074800 JXUM01074801 KQ562852 KQ560704 KXJ75006.1 KXJ81216.1 U94698 AAAB01008964 AAZX01005602 EAL39670.1 APCN01002484 ATLV01009808 KE524547 KFB35469.1 AY176050 AAO19581.1 CH477232 EAT46807.2 KX688011 ARO50443.1 AY176049 AAO19580.1 GFDF01000620 JAV13464.1 GFDF01000636 JAV13448.1 GFDF01000619 JAV13465.1 GFDF01000629 JAV13455.1 KK107021 QOIP01000011 EZA62389.1 RLU17085.1 UFQS01001766 UFQT01001766 SSX12271.1 SSX31722.1 NEVH01002150 PNF42438.1 CH933808 EDW10712.2 GANO01003040 JAB56831.1 GFDL01007245 JAV27800.1 GFDL01005067 JAV29978.1 DS232091 EDS34539.1 GGFK01007397 MBW40718.1 GGFK01007150 MBW40471.1 EDS34537.1 NEVH01027097 PNF13669.1 KJ702264 AID61418.1 UFQS01000790 UFQT01000790 SSX06908.1 SSX27252.1
BABH01001230 BABH01001231 AK289309 BAM73836.1 GU731542 ADE05592.1 KM016710 KZ149946 AID54862.1 PZC76746.1 GU731541 ADE05591.1 NWSH01002099 PCG69201.1 JX310099 AFP20610.1 KX443481 ASO98056.1 KX443480 ASO98055.1 KQ459606 KPI91147.1 KPI91146.1 KP001115 AJN91160.1 KP001116 AJN91161.1 GQ915321 ACZ97415.1 KQ460947 KPJ10553.1 KY212080 ARA91644.1 KY212081 ARA91645.1 AGBW02012935 OWR44198.1 BABH01001232 GDQN01010473 GDQN01006630 JAT80581.1 JAT84424.1 GDQN01008311 JAT82743.1 GDQN01003905 JAT87149.1 MH138217 AXP17158.1 AGBW02010730 OWR47956.1 KY212082 ARA91646.1 KPJ10552.1 GDQN01002676 JAT88378.1 GDQN01007130 JAT83924.1 KPJ10554.1 KQ459668 KPJ20511.1 KM016709 AID54861.1 MF684355 ASX93989.1 KQ459599 KPI94419.1 JX310098 AFP20609.1 AGBW02012491 OWR44987.1 KX443479 ASO98054.1 KQ461154 KPJ08442.1 GU731540 ADE05590.1 AK289310 BAM73837.1 RSAL01000062 RVE49588.1 JTDY01001897 KOB72593.1 KP001114 AJN91159.1 AXCM01012394 ODYU01008822 SOQ52761.1 UFQS01000086 UFQT01000086 SSW99091.1 SSX19473.1 GAPW01000938 JAC12660.1 JXUM01024875 JXUM01074800 JXUM01074801 KQ562852 KQ560704 KXJ75006.1 KXJ81216.1 U94698 AAAB01008964 AAZX01005602 EAL39670.1 APCN01002484 ATLV01009808 KE524547 KFB35469.1 AY176050 AAO19581.1 CH477232 EAT46807.2 KX688011 ARO50443.1 AY176049 AAO19580.1 GFDF01000620 JAV13464.1 GFDF01000636 JAV13448.1 GFDF01000619 JAV13465.1 GFDF01000629 JAV13455.1 KK107021 QOIP01000011 EZA62389.1 RLU17085.1 UFQS01001766 UFQT01001766 SSX12271.1 SSX31722.1 NEVH01002150 PNF42438.1 CH933808 EDW10712.2 GANO01003040 JAB56831.1 GFDL01007245 JAV27800.1 GFDL01005067 JAV29978.1 DS232091 EDS34539.1 GGFK01007397 MBW40718.1 GGFK01007150 MBW40471.1 EDS34537.1 NEVH01027097 PNF13669.1 KJ702264 AID61418.1 UFQS01000790 UFQT01000790 SSX06908.1 SSX27252.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000075883 UP000075920 UP000095301 UP000075885 UP000069940 UP000249989 UP000002358 UP000007062 UP000075840 UP000075903 UP000076407 UP000030765 UP000008820 UP000075900 UP000053097 UP000279307 UP000069272 UP000235965 UP000091820 UP000009192 UP000002320 UP000075880
UP000037510 UP000075883 UP000075920 UP000095301 UP000075885 UP000069940 UP000249989 UP000002358 UP000007062 UP000075840 UP000075903 UP000076407 UP000030765 UP000008820 UP000075900 UP000053097 UP000279307 UP000069272 UP000235965 UP000091820 UP000009192 UP000002320 UP000075880
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N5L2
L0N6M0
H9IVJ6
L0N731
D5L0N7
A0A068ETR9
+ More
D5L0N6 A0A2A4JB57 J7FIT7 A0A286QUH6 A0A248QEI8 A0A194PD37 A0A194PCV8 A0A0C5C533 A0A0C5C1H5 D2JLK5 A0A194QYD6 A0A1V0D9G9 A0A1V0D9G5 A0A212ERS8 H9IVJ7 A0A1E1WBR0 A0A1E1W6Z4 A0A1E1WJG1 A0A346II03 A0A212F2K7 A0A1V0D9I0 A0A194QYL5 A0A1E1WN58 A0A1E1WAC2 A0A194QZ45 A0A0N0PEW1 A0A068EWM5 A0A286MXN7 A0A194PNU6 J7FJF8 A0A212EU53 A0A248QEI9 A0A194QSH9 D5L0N5 L0N6I8 A0A3S2PF19 A0A0L7LAT5 A0A0C5C1L2 A0A182MHV7 A0A2H1WI68 A0A340TBF6 A0A1I8MP30 A0A336K3T6 A0A182PZ56 A0A023ETJ1 A0A182H3M9 O18635 A0A1S4GX15 K7J4S2 Q5TQM6 A0A182II69 A0A182UM32 A0A182XUR2 A0A182XUR0 A0A084VBX5 Q86QN3 A0A1S4GXQ5 Q17JN3 A0A1W6R7J5 Q86QN4 A0A182VHC9 A0A1L8E4A8 A0A182II71 A0A1L8E3Y1 A0A1L8E4J3 A0A1L8E4I2 A0A182S4Q4 A0A026X4P4 A0A182FZU6 A0A336MMT9 A0A2J7RNN8 A0A182PZ54 A0A1A9VZ41 B4KTY6 U5EHF6 A0A1Q3FJK3 A0A1Q3FR33 B0WTS5 A0A2M4AJC1 A0A1I8N7Q7 A0A182IJQ1 A0A2M4AI68 B0WTS3 A0A1S4JS48 A0A2J7PBF7 A0A068F5B4 A0A336MA77
D5L0N6 A0A2A4JB57 J7FIT7 A0A286QUH6 A0A248QEI8 A0A194PD37 A0A194PCV8 A0A0C5C533 A0A0C5C1H5 D2JLK5 A0A194QYD6 A0A1V0D9G9 A0A1V0D9G5 A0A212ERS8 H9IVJ7 A0A1E1WBR0 A0A1E1W6Z4 A0A1E1WJG1 A0A346II03 A0A212F2K7 A0A1V0D9I0 A0A194QYL5 A0A1E1WN58 A0A1E1WAC2 A0A194QZ45 A0A0N0PEW1 A0A068EWM5 A0A286MXN7 A0A194PNU6 J7FJF8 A0A212EU53 A0A248QEI9 A0A194QSH9 D5L0N5 L0N6I8 A0A3S2PF19 A0A0L7LAT5 A0A0C5C1L2 A0A182MHV7 A0A2H1WI68 A0A340TBF6 A0A1I8MP30 A0A336K3T6 A0A182PZ56 A0A023ETJ1 A0A182H3M9 O18635 A0A1S4GX15 K7J4S2 Q5TQM6 A0A182II69 A0A182UM32 A0A182XUR2 A0A182XUR0 A0A084VBX5 Q86QN3 A0A1S4GXQ5 Q17JN3 A0A1W6R7J5 Q86QN4 A0A182VHC9 A0A1L8E4A8 A0A182II71 A0A1L8E3Y1 A0A1L8E4J3 A0A1L8E4I2 A0A182S4Q4 A0A026X4P4 A0A182FZU6 A0A336MMT9 A0A2J7RNN8 A0A182PZ54 A0A1A9VZ41 B4KTY6 U5EHF6 A0A1Q3FJK3 A0A1Q3FR33 B0WTS5 A0A2M4AJC1 A0A1I8N7Q7 A0A182IJQ1 A0A2M4AI68 B0WTS3 A0A1S4JS48 A0A2J7PBF7 A0A068F5B4 A0A336MA77
PDB
3K9Y
E-value=1.80457e-35,
Score=375
Ontologies
GO
Topology
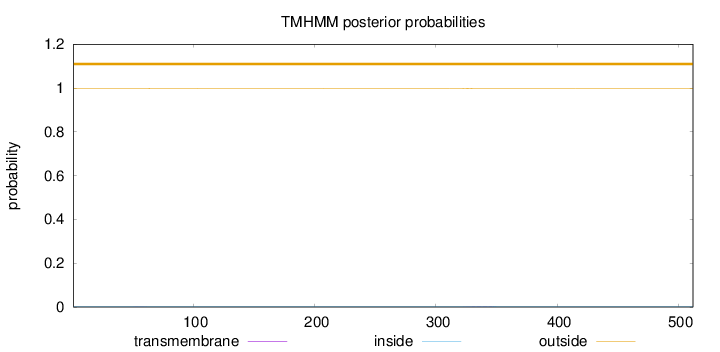
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04012
Exp number, first 60 AAs:
0.00375
Total prob of N-in:
0.00141
outside
1 - 512
Population Genetic Test Statistics
Pi
217.671875
Theta
180.838475
Tajima's D
0.389272
CLR
0.12339
CSRT
0.47727613619319
Interpretation
Uncertain
