Pre Gene Modal
BGIBMGA000866
Annotation
aspartylglucosaminidase_[Bombyx_mori]
Full name
N(4)-(Beta-N-acetylglucosaminyl)-L-asparaginase
Alternative Name
Aspartylglucosaminidase
Glycosylasparaginase
N4-(N-acetyl-beta-glucosaminyl)-L-asparagine amidase
Glycosylasparaginase
N4-(N-acetyl-beta-glucosaminyl)-L-asparagine amidase
Location in the cell
Extracellular Reliability : 1.748 Lysosomal Reliability : 1.288
Sequence
CDS
ATGAACCAAATAGGAATATTTTTAACCTATGTATTAGTTTATTTAACGAATGTTTGTTCCGAAACAAATATTCCCATAGTGATTACTACTTGGTCGTTCACAAATTCAACGGTTAAAGCTTGGGAAGTTCTAAATAACGGAGGCACGGCATTAGATGCCGTAGAGCAGGGCGCATCGGTCTGTGAAGACCAACAATGTGATGGTACTGTAGGATATGGTGGAAGTCCGGACGAAAACGGAGAAACTACATTAGATGCTCTTATTATGAATGGGAGAACTATGAATATTGGAGCTGTGGGTGGCTTGAGAAGGATCAAACATGCCATATCTGTTGCTAGACATGTATTGGACCACACAAAACATTCCTTTTTAGTTGGTGAGCTTGCCACCCAATTTGCAGTAGAAATGGGCTTTCAAGAAGAATCCCTCACTACCTCTGTATCAAAAAAACTATGGATGAAATGGCATAATGAACATCATTGTCAACCAAATTTCTGGCTGAATGTCAGTCCAGATCCCACTCGAAACTGTGGTCCATATCAGAAACACGAGTACTCTCATCATCTACTTAAGAAGAGCCATCTTAATGTGGATCGGTTCAACCATGACACGATAGGTATGGTGGCTATTGATCAACATGGTGATATTGCAGCTGGAACTTCAACAAATGGTGCTAAATATAAAATACCTGGAAGAATAGGTGATTCGCCGATTCCTGGGTCCGGCGCTTATGCTGACAATGATGTAGGCGGTGCTGCGGCGACTGGCGACGGCGATGTGATGTTGCGATTCCTTCCAAGTTTTTTAACAGTAGAAGAAATGCGCCGAGGCTCTTCACCTACTGATGCTGCTAAAACAGCAGTTGCGAGAATAGCGTCCCATTATCCGGATTTTATGGGAGCAGTTGTTGCACTGAAGAATGACGGCGAATACGGAGCGGCATGCCACGGAATAAACATGTTCCCTTATGTTGTATACGATAAAAAACATAGTTCCTATCAAATTGTTAACGTATCTTGTAGTTAA
Protein
MNQIGIFLTYVLVYLTNVCSETNIPIVITTWSFTNSTVKAWEVLNNGGTALDAVEQGASVCEDQQCDGTVGYGGSPDENGETTLDALIMNGRTMNIGAVGGLRRIKHAISVARHVLDHTKHSFLVGELATQFAVEMGFQEESLTTSVSKKLWMKWHNEHHCQPNFWLNVSPDPTRNCGPYQKHEYSHHLLKKSHLNVDRFNHDTIGMVAIDQHGDIAAGTSTNGAKYKIPGRIGDSPIPGSGAYADNDVGGAAATGDGDVMLRFLPSFLTVEEMRRGSSPTDAAKTAVARIASHYPDFMGAVVALKNDGEYGAACHGINMFPYVVYDKKHSSYQIVNVSCS
Summary
Catalytic Activity
H2O + N(4)-(beta-N-acetyl-D-glucosaminyl)-L-asparagine = H(+) + L-aspartate + N-acetyl-beta-D-glucosaminylamine
Subunit
Heterotetramer of two alpha and two beta chains or heterodimer of an alpha and a beta chain.
Similarity
Belongs to the Ntn-hydrolase family.
Keywords
Autocatalytic cleavage
Disulfide bond
Glycoprotein
Hydrolase
Lysosome
Protease
Feature
chain Glycosylasparaginase alpha chain
Uniprot
Q0PVE2
A0A2W1BNY3
A0A2A4JTX6
H9IUD6
S4PMM6
A0A194QYD1
+ More
A0A194PDP9 A0A0L7KUV7 A0A3S2LP42 A0A2H1WU24 O02467 A0A2J7Q108 A0A067RDR4 A0A2H8TU61 A0A1Q3F673 B0X0J0 A0A1W4XUG5 K7J6P3 J9JXT9 A0A232EIS5 A0A1B6KE62 A0A0K2TB28 A0A1W6EW98 A0A0C9RGS9 C1BMI4 A0A154PJC3 A0A151I7R6 A0A1Z5L6S5 A0A1D1V9Z9 A0A293MS46 A0A158NKB0 T1KBF9 A0A2A3EG38 A0A151I0K3 A0A1S3IY47 A0A224YUC8 F4W8L1 A0A1S4FM69 A0A131YXC8 E0VAH9 V5IB86 A0A087ZYB5 Q16VX6 A0A087TQ27 A0A151WIT3 E9H0E1 A0A1Y1LZF9 V5HHM1 A0A1B6G619 A0A1S7IVP7 A0A336MN77 A0A0P6CZI8 E9IBU9 A0A0P6G277 A0A1S7IVQ5 U5ELS7 A0A182U1E4 A0A164WCC5 A0A195FCC1 A0A146LSA1 A0A182KSL7 A0A0L7R0K0 A0A0P5JM61 A0A182H9B9 A0A0P5N5L1 A0A195EIS6 Q7Q079 A0A131Y1T6 A0A182G2Y0 A0A0P6C1H5 A0A182SAM8 A0A0P5PRB9 A0A0P5I204 A0A182HQ38 A0A0K8SGT2 A0A182VMH5 A0A0P5S611 A0A0P5XV16 L7M0T3 A0A0P6HPG3 A0A0P5JND4 A0A0K8U659 A0A0A1WRN7 A0A0J7KUU9 A0A2K8JWQ3 A0A182XNB7 A0A1B6MAF8 A0A1J1I8P3 A0A0P5ZNV1 D2A367 A0A0P5MCE7 A0A0P5VID2 A0A182W952 A0A3M7RFD1 A0A0C9SFJ7 F4W945 A0A195EDV6 A0A034WLU4 K7J6P2 A0A0L0BNW9
A0A194PDP9 A0A0L7KUV7 A0A3S2LP42 A0A2H1WU24 O02467 A0A2J7Q108 A0A067RDR4 A0A2H8TU61 A0A1Q3F673 B0X0J0 A0A1W4XUG5 K7J6P3 J9JXT9 A0A232EIS5 A0A1B6KE62 A0A0K2TB28 A0A1W6EW98 A0A0C9RGS9 C1BMI4 A0A154PJC3 A0A151I7R6 A0A1Z5L6S5 A0A1D1V9Z9 A0A293MS46 A0A158NKB0 T1KBF9 A0A2A3EG38 A0A151I0K3 A0A1S3IY47 A0A224YUC8 F4W8L1 A0A1S4FM69 A0A131YXC8 E0VAH9 V5IB86 A0A087ZYB5 Q16VX6 A0A087TQ27 A0A151WIT3 E9H0E1 A0A1Y1LZF9 V5HHM1 A0A1B6G619 A0A1S7IVP7 A0A336MN77 A0A0P6CZI8 E9IBU9 A0A0P6G277 A0A1S7IVQ5 U5ELS7 A0A182U1E4 A0A164WCC5 A0A195FCC1 A0A146LSA1 A0A182KSL7 A0A0L7R0K0 A0A0P5JM61 A0A182H9B9 A0A0P5N5L1 A0A195EIS6 Q7Q079 A0A131Y1T6 A0A182G2Y0 A0A0P6C1H5 A0A182SAM8 A0A0P5PRB9 A0A0P5I204 A0A182HQ38 A0A0K8SGT2 A0A182VMH5 A0A0P5S611 A0A0P5XV16 L7M0T3 A0A0P6HPG3 A0A0P5JND4 A0A0K8U659 A0A0A1WRN7 A0A0J7KUU9 A0A2K8JWQ3 A0A182XNB7 A0A1B6MAF8 A0A1J1I8P3 A0A0P5ZNV1 D2A367 A0A0P5MCE7 A0A0P5VID2 A0A182W952 A0A3M7RFD1 A0A0C9SFJ7 F4W945 A0A195EDV6 A0A034WLU4 K7J6P2 A0A0L0BNW9
EC Number
3.5.1.26
Pubmed
28756777
19121390
23622113
26354079
26227816
8877373
+ More
24845553 20075255 28648823 28528879 27649274 21347285 28797301 21719571 17510324 26830274 20566863 25765539 21292972 28004739 21282665 26823975 20966253 26483478 12364791 25576852 25830018 18362917 19820115 30375419 26131772 25348373 26108605
24845553 20075255 28648823 28528879 27649274 21347285 28797301 21719571 17510324 26830274 20566863 25765539 21292972 28004739 21282665 26823975 20966253 26483478 12364791 25576852 25830018 18362917 19820115 30375419 26131772 25348373 26108605
EMBL
DQ683192
ABG81362.1
KZ149946
PZC76769.1
NWSH01000677
PCG74860.1
+ More
BABH01001239 BABH01001240 GAIX01000096 JAA92464.1 KQ460947 KPJ10548.1 KQ459606 KPI91143.1 JTDY01005587 KOB66856.1 RSAL01000576 RVE41372.1 ODYU01010667 SOQ55924.1 S83425 NEVH01019960 PNF22271.1 KK852761 KDR17001.1 GFXV01004913 MBW16718.1 GFDL01011989 JAV23056.1 DS232239 EDS38151.1 ABLF02031677 NNAY01004159 OXU18269.1 GEBQ01030272 JAT09705.1 HACA01005315 CDW22676.1 KY563608 ARK20017.1 GBYB01006211 JAG75978.1 BT075813 ACO10237.1 KQ434912 KZC11398.1 KQ978406 KYM94138.1 GFJQ02003854 JAW03116.1 BDGG01000004 GAU98464.1 GFWV01018618 MAA43346.1 ADTU01018761 CAEY01001948 KZ288266 PBC30116.1 KQ976601 KYM79426.1 GFPF01010071 MAA21217.1 GL887908 EGI69502.1 GEDV01004714 JAP83843.1 DS235006 EEB10385.1 GANP01015872 JAB68596.1 CH477579 EAT38737.1 KK116257 KFM67216.1 KQ983082 KYQ47721.1 GL732580 EFX74790.1 GEZM01045493 JAV77780.1 GANP01011495 JAB72973.1 GECZ01011928 JAS57841.1 LC092877 BAX00386.1 UFQS01000828 UFQT01000828 SSX07095.1 SSX27438.1 GDIQ01085211 JAN09526.1 GL762137 EFZ21943.1 GDIQ01039616 JAN55121.1 LC093049 BAX00387.1 GANO01001286 JAB58585.1 LRGB01001265 KZS13141.1 KQ981693 KYN37689.1 GDHC01009319 JAQ09310.1 KQ414670 KOC64380.1 GDIQ01198109 JAK53616.1 JXUM01120177 KQ566367 KXJ70182.1 GDIQ01146990 JAL04736.1 KQ978881 KYN27764.1 AAAB01008986 EAA00283.4 GEFM01003354 JAP72442.1 JXUM01140626 KQ569095 KXJ68762.1 GDIP01006126 JAM97589.1 GDIQ01145589 JAL06137.1 GDIQ01224908 JAK26817.1 APCN01003291 GBRD01013333 JAG52493.1 GDIQ01098300 JAL53426.1 GDIP01066913 JAM36802.1 GACK01007394 JAA57640.1 GDIQ01024022 JAN70715.1 GDIQ01222306 JAK29419.1 GDHF01030273 JAI22041.1 GBXI01012568 JAD01724.1 LBMM01002883 KMQ94247.1 KY031096 ATU82847.1 GEBQ01007052 JAT32925.1 CVRI01000044 CRK96595.1 GDIP01042113 JAM61602.1 KQ971338 EFA01956.1 GDIQ01161980 JAK89745.1 GDIP01099560 JAM04155.1 REGN01003513 RNA22220.1 GBZX01000037 JAG92703.1 GL888002 EGI69227.1 KQ979039 KYN23291.1 GAKP01003625 JAC55327.1 JRES01001589 KNC21668.1
BABH01001239 BABH01001240 GAIX01000096 JAA92464.1 KQ460947 KPJ10548.1 KQ459606 KPI91143.1 JTDY01005587 KOB66856.1 RSAL01000576 RVE41372.1 ODYU01010667 SOQ55924.1 S83425 NEVH01019960 PNF22271.1 KK852761 KDR17001.1 GFXV01004913 MBW16718.1 GFDL01011989 JAV23056.1 DS232239 EDS38151.1 ABLF02031677 NNAY01004159 OXU18269.1 GEBQ01030272 JAT09705.1 HACA01005315 CDW22676.1 KY563608 ARK20017.1 GBYB01006211 JAG75978.1 BT075813 ACO10237.1 KQ434912 KZC11398.1 KQ978406 KYM94138.1 GFJQ02003854 JAW03116.1 BDGG01000004 GAU98464.1 GFWV01018618 MAA43346.1 ADTU01018761 CAEY01001948 KZ288266 PBC30116.1 KQ976601 KYM79426.1 GFPF01010071 MAA21217.1 GL887908 EGI69502.1 GEDV01004714 JAP83843.1 DS235006 EEB10385.1 GANP01015872 JAB68596.1 CH477579 EAT38737.1 KK116257 KFM67216.1 KQ983082 KYQ47721.1 GL732580 EFX74790.1 GEZM01045493 JAV77780.1 GANP01011495 JAB72973.1 GECZ01011928 JAS57841.1 LC092877 BAX00386.1 UFQS01000828 UFQT01000828 SSX07095.1 SSX27438.1 GDIQ01085211 JAN09526.1 GL762137 EFZ21943.1 GDIQ01039616 JAN55121.1 LC093049 BAX00387.1 GANO01001286 JAB58585.1 LRGB01001265 KZS13141.1 KQ981693 KYN37689.1 GDHC01009319 JAQ09310.1 KQ414670 KOC64380.1 GDIQ01198109 JAK53616.1 JXUM01120177 KQ566367 KXJ70182.1 GDIQ01146990 JAL04736.1 KQ978881 KYN27764.1 AAAB01008986 EAA00283.4 GEFM01003354 JAP72442.1 JXUM01140626 KQ569095 KXJ68762.1 GDIP01006126 JAM97589.1 GDIQ01145589 JAL06137.1 GDIQ01224908 JAK26817.1 APCN01003291 GBRD01013333 JAG52493.1 GDIQ01098300 JAL53426.1 GDIP01066913 JAM36802.1 GACK01007394 JAA57640.1 GDIQ01024022 JAN70715.1 GDIQ01222306 JAK29419.1 GDHF01030273 JAI22041.1 GBXI01012568 JAD01724.1 LBMM01002883 KMQ94247.1 KY031096 ATU82847.1 GEBQ01007052 JAT32925.1 CVRI01000044 CRK96595.1 GDIP01042113 JAM61602.1 KQ971338 EFA01956.1 GDIQ01161980 JAK89745.1 GDIP01099560 JAM04155.1 REGN01003513 RNA22220.1 GBZX01000037 JAG92703.1 GL888002 EGI69227.1 KQ979039 KYN23291.1 GAKP01003625 JAC55327.1 JRES01001589 KNC21668.1
Proteomes
UP000218220
UP000005204
UP000053240
UP000053268
UP000037510
UP000283053
+ More
UP000235965 UP000027135 UP000002320 UP000192223 UP000002358 UP000007819 UP000215335 UP000076502 UP000078542 UP000186922 UP000005205 UP000015104 UP000242457 UP000078540 UP000085678 UP000007755 UP000009046 UP000005203 UP000008820 UP000054359 UP000075809 UP000000305 UP000075902 UP000076858 UP000078541 UP000075882 UP000053825 UP000069940 UP000249989 UP000078492 UP000007062 UP000075901 UP000075840 UP000075903 UP000036403 UP000076407 UP000183832 UP000007266 UP000075920 UP000276133 UP000037069
UP000235965 UP000027135 UP000002320 UP000192223 UP000002358 UP000007819 UP000215335 UP000076502 UP000078542 UP000186922 UP000005205 UP000015104 UP000242457 UP000078540 UP000085678 UP000007755 UP000009046 UP000005203 UP000008820 UP000054359 UP000075809 UP000000305 UP000075902 UP000076858 UP000078541 UP000075882 UP000053825 UP000069940 UP000249989 UP000078492 UP000007062 UP000075901 UP000075840 UP000075903 UP000036403 UP000076407 UP000183832 UP000007266 UP000075920 UP000276133 UP000037069
SUPFAM
SSF56235
SSF56235
ProteinModelPortal
Q0PVE2
A0A2W1BNY3
A0A2A4JTX6
H9IUD6
S4PMM6
A0A194QYD1
+ More
A0A194PDP9 A0A0L7KUV7 A0A3S2LP42 A0A2H1WU24 O02467 A0A2J7Q108 A0A067RDR4 A0A2H8TU61 A0A1Q3F673 B0X0J0 A0A1W4XUG5 K7J6P3 J9JXT9 A0A232EIS5 A0A1B6KE62 A0A0K2TB28 A0A1W6EW98 A0A0C9RGS9 C1BMI4 A0A154PJC3 A0A151I7R6 A0A1Z5L6S5 A0A1D1V9Z9 A0A293MS46 A0A158NKB0 T1KBF9 A0A2A3EG38 A0A151I0K3 A0A1S3IY47 A0A224YUC8 F4W8L1 A0A1S4FM69 A0A131YXC8 E0VAH9 V5IB86 A0A087ZYB5 Q16VX6 A0A087TQ27 A0A151WIT3 E9H0E1 A0A1Y1LZF9 V5HHM1 A0A1B6G619 A0A1S7IVP7 A0A336MN77 A0A0P6CZI8 E9IBU9 A0A0P6G277 A0A1S7IVQ5 U5ELS7 A0A182U1E4 A0A164WCC5 A0A195FCC1 A0A146LSA1 A0A182KSL7 A0A0L7R0K0 A0A0P5JM61 A0A182H9B9 A0A0P5N5L1 A0A195EIS6 Q7Q079 A0A131Y1T6 A0A182G2Y0 A0A0P6C1H5 A0A182SAM8 A0A0P5PRB9 A0A0P5I204 A0A182HQ38 A0A0K8SGT2 A0A182VMH5 A0A0P5S611 A0A0P5XV16 L7M0T3 A0A0P6HPG3 A0A0P5JND4 A0A0K8U659 A0A0A1WRN7 A0A0J7KUU9 A0A2K8JWQ3 A0A182XNB7 A0A1B6MAF8 A0A1J1I8P3 A0A0P5ZNV1 D2A367 A0A0P5MCE7 A0A0P5VID2 A0A182W952 A0A3M7RFD1 A0A0C9SFJ7 F4W945 A0A195EDV6 A0A034WLU4 K7J6P2 A0A0L0BNW9
A0A194PDP9 A0A0L7KUV7 A0A3S2LP42 A0A2H1WU24 O02467 A0A2J7Q108 A0A067RDR4 A0A2H8TU61 A0A1Q3F673 B0X0J0 A0A1W4XUG5 K7J6P3 J9JXT9 A0A232EIS5 A0A1B6KE62 A0A0K2TB28 A0A1W6EW98 A0A0C9RGS9 C1BMI4 A0A154PJC3 A0A151I7R6 A0A1Z5L6S5 A0A1D1V9Z9 A0A293MS46 A0A158NKB0 T1KBF9 A0A2A3EG38 A0A151I0K3 A0A1S3IY47 A0A224YUC8 F4W8L1 A0A1S4FM69 A0A131YXC8 E0VAH9 V5IB86 A0A087ZYB5 Q16VX6 A0A087TQ27 A0A151WIT3 E9H0E1 A0A1Y1LZF9 V5HHM1 A0A1B6G619 A0A1S7IVP7 A0A336MN77 A0A0P6CZI8 E9IBU9 A0A0P6G277 A0A1S7IVQ5 U5ELS7 A0A182U1E4 A0A164WCC5 A0A195FCC1 A0A146LSA1 A0A182KSL7 A0A0L7R0K0 A0A0P5JM61 A0A182H9B9 A0A0P5N5L1 A0A195EIS6 Q7Q079 A0A131Y1T6 A0A182G2Y0 A0A0P6C1H5 A0A182SAM8 A0A0P5PRB9 A0A0P5I204 A0A182HQ38 A0A0K8SGT2 A0A182VMH5 A0A0P5S611 A0A0P5XV16 L7M0T3 A0A0P6HPG3 A0A0P5JND4 A0A0K8U659 A0A0A1WRN7 A0A0J7KUU9 A0A2K8JWQ3 A0A182XNB7 A0A1B6MAF8 A0A1J1I8P3 A0A0P5ZNV1 D2A367 A0A0P5MCE7 A0A0P5VID2 A0A182W952 A0A3M7RFD1 A0A0C9SFJ7 F4W945 A0A195EDV6 A0A034WLU4 K7J6P2 A0A0L0BNW9
PDB
1APZ
E-value=6.34926e-54,
Score=532
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Lysosome
SignalP
Position: 1 - 20,
Likelihood: 0.965139
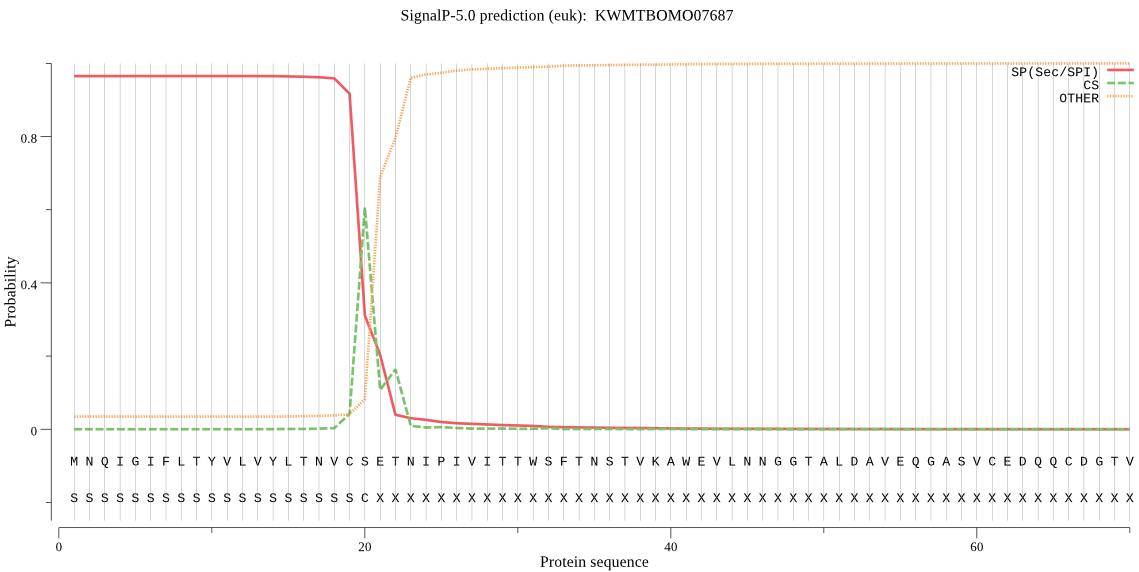
Length:
341
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.57914000000001
Exp number, first 60 AAs:
3.57087
Total prob of N-in:
0.15804
outside
1 - 341
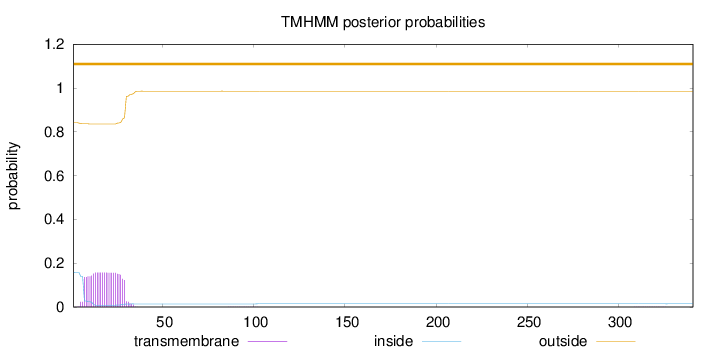
Population Genetic Test Statistics
Pi
25.600469
Theta
155.805145
Tajima's D
-1.684414
CLR
1.987734
CSRT
0.0380980950952452
Interpretation
Uncertain
